README.md
In RuzhangZhao/Immigrate: Iterative Max-Min Entropy Margin-Maximization with Interaction Terms for Feature Selection
Immigrate : A Margin-Based Feature Selection Method with Interaction Terms
: A Margin-Based Feature Selection Method with Interaction Terms
This project is the R code for IMMIGRATE method (Iterative Max-MIn entropy marGin-maximization with inteRAction TErms algorithm, IMMIGRATE, henceforth).
IMMIGRATE is a hypothesis-margin based feature selection method with interaction terms. For more details, please refer to the paper (published version , arXiv).
Based on large hypothesis-margin principle, this package performs some feature selection methods:
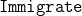 (Iterative Max-Min Entropy Margin-Maximization with Interaction Terms Algorithm);
(Iterative Max-Min Entropy Margin-Maximization with Interaction Terms Algorithm);
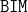 (Booster version of IMMIGRATE);
(Booster version of IMMIGRATE);
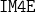 (Iterative Margin-Maximization under Max-Min Entropy Algorithm);
(Iterative Margin-Maximization under Max-Min Entropy Algorithm);
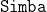 (Iterative Search Margin Based Algorithm);
(Iterative Search Margin Based Algorithm);
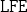 (Local Feature Extraction Algorithm).
(Local Feature Extraction Algorithm).
This package also performs prediction for the above feature selection methods.
Installation
if(!require(Immigrate)){
install.packages("Immigrate")
}
library(Immigrate)
Our R package Immigrate is released on CRAN. Please use this link https://cran.r-project.org/package=Immigrate to refer to the package page on CRAN.
Or people can install from Github.
if(!require(devtools)){
install.packages("devtools")
}
devtools::install_github("RuzhangZhao/Immigrate")
Check the package version.
packageVersion("Immigrate")
Implementation Demo
We first provide implementation demo for the method IMMIGRATE, and then, we compare the performance of IMMIGRATE with other popular methods.
The data we use in this demo is from UCI Machine Learning Repository (here).
Dataset name: Oxford Parkinson's Disease Detection Dataset.
We have uploaded the Parkinson's Disease Dataset in R package Immigrate and named it as park. The Parkinson's Disease Detection Dataset is loaded easily as follows.
data("park")
dim(park$xx) # 194 22
length(park$yy) # 194
Implementation of IMMIGRATE
The default implementation of IMMIGRATE needs the training explanatory data matrix (sample size $\times$ number of feature) and training labels.
Using all the data as training data, we have
demo_Immigrate<-Immigrate(park$xx,park$yy)
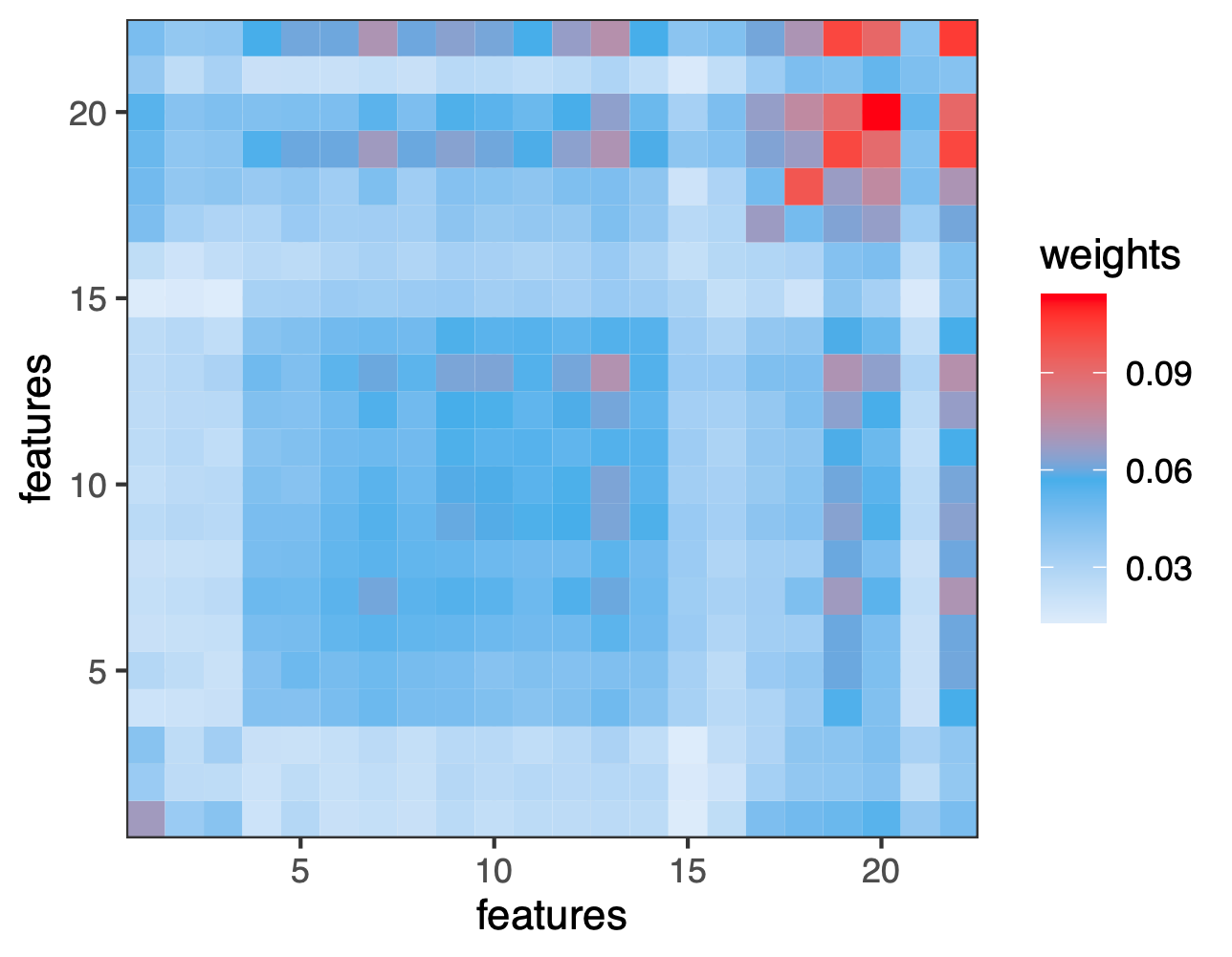
To visualize the results, we show an interesting heat map from the weight matrix obtained here.
if(!require(ggplot2)){
install.packages("ggplot2")
}
if(!require(reshape2)){
install.packages("reshape2")
}
library(ggplot2)
library(reshape2)
demo_w_melt<-melt(demo_Immigrate$w)
demo_heat_map <- ggplot(data = demo_w_melt) +
geom_tile(aes(x = Var1, y = Var2, fill = value))+
theme_bw()+
scale_fill_gradient2("weights",midpoint = max(demo_w_melt$value)/2,
low = "white",
mid = "steelblue2",
high = "red")+
theme(panel.grid.minor = element_line(size=1))+
scale_x_continuous( expand = c(0, 0))+
scale_y_continuous( expand = c(0, 0))+
labs(x = "features", y= "features")
One can refer to the CRAN page of Immigrate for details.
IMMIGRATE vs Other Methods
We use a demo to compare the performance of IMMIGRATE with Generalized Linear Model (GLM).
We use 70%/30% data as training/test data.
if(!require(caret)){
install.packages("caret")
}
library(caret)
# set seed for random data partition
set.seed(2020)
# 70% data as training data
# 30% data as test data
partition_index<-createDataPartition(park$yy,p=0.7)
train_xx<-park$xx[partition_index$Resample1,]
test_xx<-park$xx[-partition_index$Resample1,]
train_yy<-park$yy[partition_index$Resample1]
test_yy<-park$yy[-partition_index$Resample1]
We use logistic regression for GLM.
# glm training
res_glm<-glm(as.factor(train_yy)~.,
data = train_df,family = "binomial")
# glm prediction
pred_res_glm<-predict(res_glm,
newdata = data.frame(test_xx),
type = "response")
pred_res_glm<-ifelse(pred_res_glm>.5,1,0)
sum(pred_res_glm == test_yy)/length(test_yy)
We use default training setting from IMMIGRATE.
# IMMIGRATE training
res_Immigrate<-Immigrate(train_xx,train_yy)
# IMMIGRATE prediction
pred_res_Immigrate<-predict(res_Immigrate,
xx = train_xx,
yy = train_yy,
newx = test_xx,
type = "class")
sum(pred_res_glm == test_yy)/length(test_yy)
The accuracy on test data is 0.793 (GLM) vs 0.914 (IMMIGRATE).
Note
- Based on our experiments, the maximal iteration number in Immigrate does not need to be set too large.
- The performance of IMMIGRATE also depends on the choice of
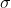 ( sig in function Immigrate).
( sig in function Immigrate).
- The default weight pruning strategy is not removing small weights. One can choose to remove small weights by calling removesmall = TRUE in function Immigrate.
- The default initial weight matrix is diagonal matrix. One can choose to use random initialization by calling randomw0 = TRUE in function Immigrate.
Authors
Ruzhang Zhao, Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD 21205, USA
Pengyu Hong, Department of Computer Science, Brandeis University, Waltham, MA 02453, USA
Jun S. Liu, Department of Statistics, Harvard University, Cambridge, MA 02138, USA
Reference
Please use the link https://www.mdpi.com/1099-4300/22/3/291 for our paper: IMMIGRATE: A Margin-Based Feature Selection Method with Interaction Terms.
Please use the link https://cran.r-project.org/package=Immigrate for the R package.
We also implement the following three hypothesis-margin based methods in this R package.
IM4E: Bei, Yuanzhe, and Pengyu Hong. "Maximizing margin quality and quantity." 2015 IEEE 25th International Workshop on Machine Learning for Signal Processing (MLSP). IEEE, 2015.
Simba: Gilad-Bachrach, Ran, Amir Navot, and Naftali Tishby. "Margin based feature selection-theory and algorithms." Proceedings of the twenty-first international conference on Machine learning. 2004.
LFE: Sun, Yijun, and Dapeng Wu. "A relief based feature extraction algorithm." Proceedings of the 2008 SIAM International Conference on Data Mining. Society for Industrial and Applied Mathematics, 2008.
RuzhangZhao/Immigrate documentation built on May 24, 2020, 5:38 a.m.
Immigrate : A Margin-Based Feature Selection Method with Interaction Terms
: A Margin-Based Feature Selection Method with Interaction Terms
This project is the R code for IMMIGRATE method (Iterative Max-MIn entropy marGin-maximization with inteRAction TErms algorithm, IMMIGRATE, henceforth).
IMMIGRATE is a hypothesis-margin based feature selection method with interaction terms. For more details, please refer to the paper (published version , arXiv).
Based on large hypothesis-margin principle, this package performs some feature selection methods:
(Iterative Max-Min Entropy Margin-Maximization with Interaction Terms Algorithm);
(Booster version of IMMIGRATE);
(Iterative Margin-Maximization under Max-Min Entropy Algorithm);
(Iterative Search Margin Based Algorithm);
(Local Feature Extraction Algorithm).
This package also performs prediction for the above feature selection methods.
Installation
if(!require(Immigrate)){
install.packages("Immigrate")
}
library(Immigrate)
Our R package Immigrate is released on CRAN. Please use this link https://cran.r-project.org/package=Immigrate to refer to the package page on CRAN.
Or people can install from Github.
if(!require(devtools)){
install.packages("devtools")
}
devtools::install_github("RuzhangZhao/Immigrate")
Check the package version.
packageVersion("Immigrate")
Implementation Demo
We first provide implementation demo for the method IMMIGRATE, and then, we compare the performance of IMMIGRATE with other popular methods.
The data we use in this demo is from UCI Machine Learning Repository (here).
Dataset name: Oxford Parkinson's Disease Detection Dataset.
We have uploaded the Parkinson's Disease Dataset in R package Immigrate and named it as park. The Parkinson's Disease Detection Dataset is loaded easily as follows.
data("park")
dim(park$xx) # 194 22
length(park$yy) # 194
Implementation of IMMIGRATE
The default implementation of IMMIGRATE needs the training explanatory data matrix (sample size $\times$ number of feature) and training labels.
Using all the data as training data, we have
demo_Immigrate<-Immigrate(park$xx,park$yy)
To visualize the results, we show an interesting heat map from the weight matrix obtained here.
if(!require(ggplot2)){
install.packages("ggplot2")
}
if(!require(reshape2)){
install.packages("reshape2")
}
library(ggplot2)
library(reshape2)
demo_w_melt<-melt(demo_Immigrate$w)
demo_heat_map <- ggplot(data = demo_w_melt) +
geom_tile(aes(x = Var1, y = Var2, fill = value))+
theme_bw()+
scale_fill_gradient2("weights",midpoint = max(demo_w_melt$value)/2,
low = "white",
mid = "steelblue2",
high = "red")+
theme(panel.grid.minor = element_line(size=1))+
scale_x_continuous( expand = c(0, 0))+
scale_y_continuous( expand = c(0, 0))+
labs(x = "features", y= "features")

One can refer to the CRAN page of Immigrate for details.
IMMIGRATE vs Other Methods
We use a demo to compare the performance of IMMIGRATE with Generalized Linear Model (GLM).
We use 70%/30% data as training/test data.
if(!require(caret)){
install.packages("caret")
}
library(caret)
# set seed for random data partition
set.seed(2020)
# 70% data as training data
# 30% data as test data
partition_index<-createDataPartition(park$yy,p=0.7)
train_xx<-park$xx[partition_index$Resample1,]
test_xx<-park$xx[-partition_index$Resample1,]
train_yy<-park$yy[partition_index$Resample1]
test_yy<-park$yy[-partition_index$Resample1]
We use logistic regression for GLM.
# glm training
res_glm<-glm(as.factor(train_yy)~.,
data = train_df,family = "binomial")
# glm prediction
pred_res_glm<-predict(res_glm,
newdata = data.frame(test_xx),
type = "response")
pred_res_glm<-ifelse(pred_res_glm>.5,1,0)
sum(pred_res_glm == test_yy)/length(test_yy)
We use default training setting from IMMIGRATE.
# IMMIGRATE training
res_Immigrate<-Immigrate(train_xx,train_yy)
# IMMIGRATE prediction
pred_res_Immigrate<-predict(res_Immigrate,
xx = train_xx,
yy = train_yy,
newx = test_xx,
type = "class")
sum(pred_res_glm == test_yy)/length(test_yy)
The accuracy on test data is 0.793 (GLM) vs 0.914 (IMMIGRATE).
Note
- Based on our experiments, the maximal iteration number in Immigrate does not need to be set too large.
- The performance of IMMIGRATE also depends on the choice of
( sig in function Immigrate).
- The default weight pruning strategy is not removing small weights. One can choose to remove small weights by calling removesmall = TRUE in function Immigrate.
- The default initial weight matrix is diagonal matrix. One can choose to use random initialization by calling randomw0 = TRUE in function Immigrate.
Authors
Ruzhang Zhao, Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD 21205, USA
Pengyu Hong, Department of Computer Science, Brandeis University, Waltham, MA 02453, USA
Jun S. Liu, Department of Statistics, Harvard University, Cambridge, MA 02138, USA
Reference
Please use the link https://www.mdpi.com/1099-4300/22/3/291 for our paper: IMMIGRATE: A Margin-Based Feature Selection Method with Interaction Terms.
Please use the link https://cran.r-project.org/package=Immigrate for the R package.
We also implement the following three hypothesis-margin based methods in this R package.
IM4E: Bei, Yuanzhe, and Pengyu Hong. "Maximizing margin quality and quantity." 2015 IEEE 25th International Workshop on Machine Learning for Signal Processing (MLSP). IEEE, 2015.
Simba: Gilad-Bachrach, Ran, Amir Navot, and Naftali Tishby. "Margin based feature selection-theory and algorithms." Proceedings of the twenty-first international conference on Machine learning. 2004.
LFE: Sun, Yijun, and Dapeng Wu. "A relief based feature extraction algorithm." Proceedings of the 2008 SIAM International Conference on Data Mining. Society for Industrial and Applied Mathematics, 2008.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
