README.md
In TiagoOlivoto/WAASB: Multi Environment Trials Analysis
metan 
metan (multi-environment trials analysis) provides
useful functions for analyzing multi-environment trial data using
parametric and non-parametric methods. The package will help you to:
- Inspect
data for possible common errors;
- Manipulate rows and
columns;
- Manipulate numbers and
strings;
- Manipulate
NAs and
0s;
- Compute descriptive
statistics;
- Compute
within-environment
and joint-analysis of
variance;
- Compute AMMI
analysis
with prediction considering different numbers of interaction principal
component axes;
- Compute AMMI-based stability
indexes;
- Compute GGE biplot
analysis;
- Compute GT
and GYT
biplot analysis;
- Compute BLUP-based stability
indexes;
- Compute variance components and genetic parameters in single
environment
and
multi-environment
trials using mixed-effect models;
- Perform cross-validation procedures for
AMMI-family
and
BLUP
models;
- Compute parametric and nonparametric stability
statistics;
- Implement biometrical
models.
Installation
Install the released version of metan from
CRAN with:
install.packages("metan")
Or install the development version from
GitHub with:
devtools::install_github("TiagoOlivoto/metan")
# To build the HTML vignette use
devtools::install_github("TiagoOlivoto/metan", build_vignettes = TRUE)
Note: If you are a Windows user, you should also first download and
install the latest version of
Rtools.
For the latest release notes on this development version, see the NEWS
file.
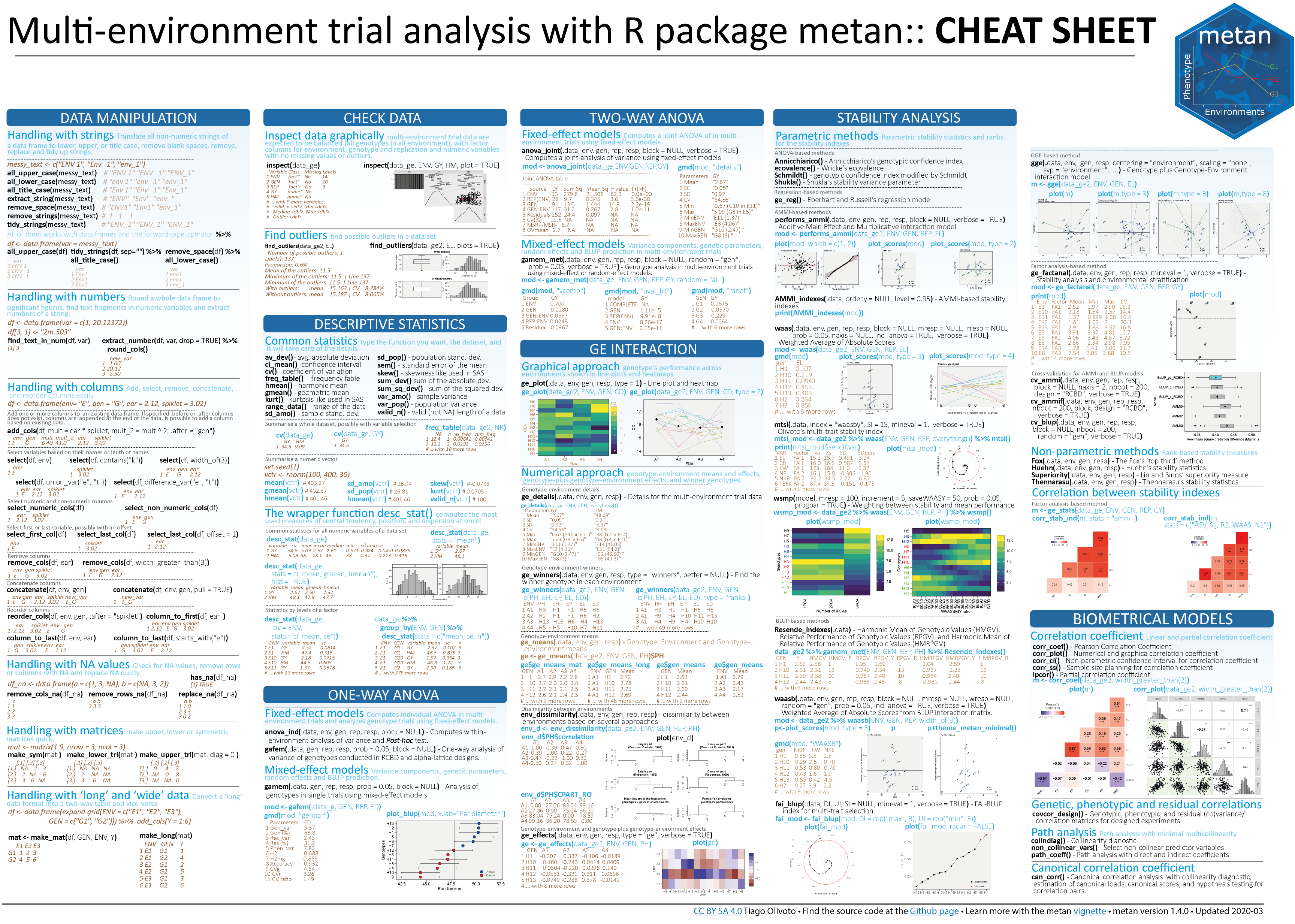
Cheatsheet
Getting started
metan offers a set of functions that can be used to manipulate,
summarize, analyze and plot typical multi-environment trial data. Maybe,
one of the first functions users should use would be
inspect().
Here, we will inspect the example dataset data_ge that contains data
on two variables assessed in 10 genotypes growing in 14 environments.
library(metan)
inspect(data_ge, plot = TRUE)
# # A tibble: 5 × 10
# Variable Class Missing Levels Valid_n Min Median Max Outlier Text
# <chr> <chr> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <lgl>
# 1 ENV factor No 14 420 NA NA NA NA NA
# 2 GEN factor No 10 420 NA NA NA NA NA
# 3 REP factor No 3 420 NA NA NA NA NA
# 4 GY numeric No - 420 0.67 2.61 5.09 0 NA
# 5 HM numeric No - 420 38 48 58 0 NA

No issues while inspecting the data. If any issue is given here (like
outliers, missing values, etc.) consider using
find_outliers()
to find possible outliers in the data set or any metan’s data
manipulation tool such as
remove_rows_na()
to remove rows with NA values,
replace_zero()
to replace 0’s with NA,
as_factor()
to convert desired columns to factor,
find_text_in_num()
to find text fragments in columns assumed to be numeric, or even
tidy_strings()
to tidy up strings.
Descriptive statistics
metan provides a set of
functions
to compute descriptive statistics. The easiest way to do that is by
using
desc_stat().
desc_stat(data_ge2)
# # A tibble: 15 × 10
# variable cv max mean median min sd.amo se ci.t n.valid
# <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
# 1 CD 7.34 18.6 16.0 16 12.9 1.17 0.0939 0.186 156
# 2 CDED 5.71 0.694 0.586 0.588 0.495 0.0334 0.0027 0.0053 156
# 3 CL 7.95 34.7 29.0 28.7 23.5 2.31 0.185 0.365 156
# 4 CW 25.2 38.5 24.8 24.5 11.1 6.26 0.501 0.99 156
# 5 ED 5.58 54.9 49.5 49.9 43.5 2.76 0.221 0.437 156
# 6 EH 21.2 1.88 1.34 1.41 0.752 0.284 0.0228 0.045 156
# 7 EL 8.28 17.9 15.2 15.1 11.5 1.26 0.101 0.199 156
# 8 EP 10.5 0.660 0.537 0.544 0.386 0.0564 0.0045 0.0089 156
# 9 KW 18.9 251. 173. 175. 106. 32.8 2.62 5.18 156
# 10 NKE 14.2 697. 512. 509. 332. 72.6 5.82 11.5 156
# 11 NKR 10.7 42 32.2 32 23.2 3.47 0.277 0.548 156
# 12 NR 10.2 21.2 16.1 16 12.4 1.64 0.131 0.259 156
# 13 PERK 2.17 91.8 87.4 87.5 81.2 1.90 0.152 0.300 156
# 14 PH 13.4 3.04 2.48 2.52 1.71 0.334 0.0267 0.0528 156
# 15 TKW 13.9 452. 339. 342. 218. 47.1 3.77 7.44 156
AMMI model
Fitting the model
The AMMI model is fitted with the function
performs_ammi().
To analyze multiple variables at once we can use a comma-separated
vector of unquoted variable names, or use any select helper in the
argument resp. Here, using everything() we apply the function to all
numeric variables in the data. For more details, see the complete
vignette.
model <- performs_ammi(data_ge,
env = ENV,
gen = GEN,
rep = REP,
resp = everything(),
verbose = FALSE)
# Significance of IPCAs
get_model_data(model, "ipca_pval")
# Class of the model: performs_ammi
# Variable extracted: Pr(>F)
# # A tibble: 9 × 4
# PC DF GY HM
# <chr> <dbl> <dbl> <dbl>
# 1 PC1 21 0 0
# 2 PC2 19 0 0
# 3 PC3 17 0.0014 0.0021
# 4 PC4 15 0.0096 0.0218
# 5 PC5 13 0.318 0.0377
# 6 PC6 11 0.561 0.041
# 7 PC7 9 0.754 0.0633
# 8 PC8 7 0.804 0.232
# 9 PC9 5 0.934 0.944
Biplots
The well-known AMMI1 and AMMI2 biplots can be created with
plot_scores().
Note that since
performs_ammi
allows analyzing multiple variables at once, e.g.,
resp = c(v1, v2, ...), the output model is a list, in this case with
two elements (GY and HM). By default, the biplots are created for the
first variable of the model. To choose another variable use the argument
var (e.g., var = "HM").
a <- plot_scores(model)
b <- plot_scores(model,
type = 2, # AMMI 2 biplot
polygon = TRUE, # show a polygon
highlight = c("G4", "G5", "G6"), #highlight genotypes
col.alpha.env = 0.5, # alpha for environments
col.alpha.gen = 0, # remove the other genotypes
col.env = "gray", # color for environment point
col.segm.env = "gray", # color for environment segment
plot_theme = theme_metan_minimal()) # theme
arrange_ggplot(a, b, tag_levels = "a")

GGE model
The GGE model is fitted with the function
gge(). For
more details, see the complete
vignette.
model <- gge(data_ge, ENV, GEN, GY)
model2 <- gge(data_ge, ENV, GEN, GY, svp = "genotype")
model3 <- gge(data_ge, ENV, GEN, GY, svp = "symmetrical")
a <- plot(model)
b <- plot(model2, type = 8)
c <- plot(model2,
type = 2,
col.gen = "black",
col.env = "gray70",
axis.expand = 1.5,
plot_theme = theme_metan_minimal())
arrange_ggplot(a, b, c, tag_levels = "a")

BLUP model
Linear-mixed effect models to predict the response variable in METs are
fitted using the function
gamem_met().
Here we will obtain the predicted means for genotypes in the variables
GY and HM. For more details, see the complete
vignette.
model2 <-
gamem_met(data_ge,
env = ENV,
gen = GEN,
rep = REP,
resp = everything())
# Evaluating trait GY |====================== | 50% 00:00:01 Evaluating trait HM |============================================| 100% 00:00:02
# Method: REML/BLUP
# Random effects: GEN, GEN:ENV
# Fixed effects: ENV, REP(ENV)
# Denominador DF: Satterthwaite's method
# ---------------------------------------------------------------------------
# P-values for Likelihood Ratio Test of the analyzed traits
# ---------------------------------------------------------------------------
# model GY HM
# COMPLETE NA NA
# GEN 1.11e-05 5.07e-03
# GEN:ENV 2.15e-11 2.27e-15
# ---------------------------------------------------------------------------
# All variables with significant (p < 0.05) genotype-vs-environment interaction
# Get the variance components
get_model_data(model2, what = "vcomp")
# Class of the model: waasb
# Variable extracted: vcomp
# # A tibble: 3 × 3
# Group GY HM
# <chr> <dbl> <dbl>
# 1 GEN 0.0280 0.490
# 2 GEN:ENV 0.0567 2.19
# 3 Residual 0.0967 2.84
Plotting the BLUPs for genotypes
To produce a plot with the predicted means, use the function
plot_blup().
a <- plot_blup(model2)
b <- plot_blup(model2,
prob = 0.2,
col.shape = c("gray20", "gray80"),
invert = TRUE)
arrange_ggplot(a, b, tag_levels = "a")

Computing parametric and non-parametric stability indexes
The easiest way to compute parametric and non-parametric stability
indexes in metan is by using the function
ge_stats().
It is a wrapper function around a lot of specific functions for
stability indexes. To get the results into a “ready-to-read” file, use
get_model_data()
or its shortcut
gmd().
stats <- ge_stats(data_ge, ENV, GEN, REP, GY)
# Evaluating trait GY |============================================| 100% 00:00:08
get_model_data(stats)
# Class of the model: ge_stats
# Variable extracted: stats
# # A tibble: 10 × 44
# var GEN Y CV ACV POLAR Var Shukla Wi_g Wi_f Wi_u Ecoval
# <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
# 1 GY G1 2.60 35.2 34.1 0.0298 10.9 0.0280 84.4 89.2 81.1 1.22
# 2 GY G10 2.47 42.3 38.6 0.136 14.2 0.244 59.2 64.6 54.4 7.96
# 3 GY G2 2.74 34.0 35.2 0.0570 11.3 0.0861 82.8 95.3 75.6 3.03
# 4 GY G3 2.96 29.9 33.8 0.0216 10.1 0.0121 104. 99.7 107. 0.725
# 5 GY G4 2.64 31.4 31.0 -0.0537 8.93 0.0640 85.9 79.5 91.9 2.34
# 6 GY G5 2.54 30.6 28.8 -0.119 7.82 0.0480 82.7 82.2 82.4 1.84
# 7 GY G6 2.53 29.7 27.8 -0.147 7.34 0.0468 83.0 83.7 81.8 1.81
# 8 GY G7 2.74 27.4 28.3 -0.133 7.33 0.122 83.9 77.6 93.4 4.16
# 9 GY G8 3.00 30.4 35.1 0.0531 10.8 0.0712 98.8 90.5 107. 2.57
# 10 GY G9 2.51 42.4 39.4 0.154 14.7 0.167 68.8 68.9 70.3 5.56
# # … with 32 more variables: bij <dbl>, Sij <dbl>, R2 <dbl>, ASTAB <dbl>,
# # ASI <dbl>, ASV <dbl>, AVAMGE <dbl>, DA <dbl>, DZ <dbl>, EV <dbl>, FA <dbl>,
# # MASI <dbl>, MASV <dbl>, SIPC <dbl>, ZA <dbl>, WAAS <dbl>, WAASB <dbl>,
# # HMGV <dbl>, RPGV <dbl>, HMRPGV <dbl>, Pi_a <dbl>, Pi_f <dbl>, Pi_u <dbl>,
# # Gai <dbl>, S1 <dbl>, S2 <dbl>, S3 <dbl>, S6 <dbl>, N1 <dbl>, N2 <dbl>,
# # N3 <dbl>, N4 <dbl>
Citation
citation("metan")
Please, support this project by citing it in your publications!
Olivoto, T., and Lúcio, A.D. (2020). metan: an R package for
multi-environment trial analysis. Methods Ecol Evol. 11:783-789
doi:10.1111/2041-210X.13384
A BibTeX entry for LaTeX users is
@Article{Olivoto2020,
author = {Tiago Olivoto and Alessandro Dal'Col L{'{u}}cio},
title = {metan: an R package for multi-environment trial analysis},
journal = {Methods in Ecology and Evolution},
volume = {11},
number = {6},
pages = {783-789},
year = {2020},
doi = {10.1111/2041-210X.13384},
}
Getting help
-
If you encounter a clear bug, please file a minimal reproducible
example on github
-
Suggestions and criticisms to improve the quality and usability of the
package are welcome!
TiagoOlivoto/WAASB documentation built on March 20, 2024, 4:18 p.m.
metan 
metan (multi-environment trials analysis) provides
useful functions for analyzing multi-environment trial data using
parametric and non-parametric methods. The package will help you to:
- Inspect data for possible common errors;
- Manipulate rows and columns;
- Manipulate numbers and strings;
- Manipulate
NAs and0s; - Compute descriptive statistics;
- Compute within-environment and joint-analysis of variance;
- Compute AMMI analysis with prediction considering different numbers of interaction principal component axes;
- Compute AMMI-based stability indexes;
- Compute GGE biplot analysis;
- Compute GT and GYT biplot analysis;
- Compute BLUP-based stability indexes;
- Compute variance components and genetic parameters in single environment and multi-environment trials using mixed-effect models;
- Perform cross-validation procedures for AMMI-family and BLUP models;
- Compute parametric and nonparametric stability statistics;
- Implement biometrical models.
Installation
Install the released version of metan from
CRAN with:
install.packages("metan")
Or install the development version from GitHub with:
devtools::install_github("TiagoOlivoto/metan")
# To build the HTML vignette use
devtools::install_github("TiagoOlivoto/metan", build_vignettes = TRUE)
Note: If you are a Windows user, you should also first download and install the latest version of Rtools.
For the latest release notes on this development version, see the NEWS file.
Cheatsheet
Getting started
metan offers a set of functions that can be used to manipulate,
summarize, analyze and plot typical multi-environment trial data. Maybe,
one of the first functions users should use would be
inspect().
Here, we will inspect the example dataset data_ge that contains data
on two variables assessed in 10 genotypes growing in 14 environments.
library(metan)
inspect(data_ge, plot = TRUE)
# # A tibble: 5 × 10
# Variable Class Missing Levels Valid_n Min Median Max Outlier Text
# <chr> <chr> <chr> <chr> <int> <dbl> <dbl> <dbl> <dbl> <lgl>
# 1 ENV factor No 14 420 NA NA NA NA NA
# 2 GEN factor No 10 420 NA NA NA NA NA
# 3 REP factor No 3 420 NA NA NA NA NA
# 4 GY numeric No - 420 0.67 2.61 5.09 0 NA
# 5 HM numeric No - 420 38 48 58 0 NA

No issues while inspecting the data. If any issue is given here (like
outliers, missing values, etc.) consider using
find_outliers()
to find possible outliers in the data set or any metan’s data
manipulation tool such as
remove_rows_na()
to remove rows with NA values,
replace_zero()
to replace 0’s with NA,
as_factor()
to convert desired columns to factor,
find_text_in_num()
to find text fragments in columns assumed to be numeric, or even
tidy_strings()
to tidy up strings.
Descriptive statistics
metan provides a set of
functions
to compute descriptive statistics. The easiest way to do that is by
using
desc_stat().
desc_stat(data_ge2)
# # A tibble: 15 × 10
# variable cv max mean median min sd.amo se ci.t n.valid
# <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
# 1 CD 7.34 18.6 16.0 16 12.9 1.17 0.0939 0.186 156
# 2 CDED 5.71 0.694 0.586 0.588 0.495 0.0334 0.0027 0.0053 156
# 3 CL 7.95 34.7 29.0 28.7 23.5 2.31 0.185 0.365 156
# 4 CW 25.2 38.5 24.8 24.5 11.1 6.26 0.501 0.99 156
# 5 ED 5.58 54.9 49.5 49.9 43.5 2.76 0.221 0.437 156
# 6 EH 21.2 1.88 1.34 1.41 0.752 0.284 0.0228 0.045 156
# 7 EL 8.28 17.9 15.2 15.1 11.5 1.26 0.101 0.199 156
# 8 EP 10.5 0.660 0.537 0.544 0.386 0.0564 0.0045 0.0089 156
# 9 KW 18.9 251. 173. 175. 106. 32.8 2.62 5.18 156
# 10 NKE 14.2 697. 512. 509. 332. 72.6 5.82 11.5 156
# 11 NKR 10.7 42 32.2 32 23.2 3.47 0.277 0.548 156
# 12 NR 10.2 21.2 16.1 16 12.4 1.64 0.131 0.259 156
# 13 PERK 2.17 91.8 87.4 87.5 81.2 1.90 0.152 0.300 156
# 14 PH 13.4 3.04 2.48 2.52 1.71 0.334 0.0267 0.0528 156
# 15 TKW 13.9 452. 339. 342. 218. 47.1 3.77 7.44 156
AMMI model
Fitting the model
The AMMI model is fitted with the function
performs_ammi().
To analyze multiple variables at once we can use a comma-separated
vector of unquoted variable names, or use any select helper in the
argument resp. Here, using everything() we apply the function to all
numeric variables in the data. For more details, see the complete
vignette.
model <- performs_ammi(data_ge,
env = ENV,
gen = GEN,
rep = REP,
resp = everything(),
verbose = FALSE)
# Significance of IPCAs
get_model_data(model, "ipca_pval")
# Class of the model: performs_ammi
# Variable extracted: Pr(>F)
# # A tibble: 9 × 4
# PC DF GY HM
# <chr> <dbl> <dbl> <dbl>
# 1 PC1 21 0 0
# 2 PC2 19 0 0
# 3 PC3 17 0.0014 0.0021
# 4 PC4 15 0.0096 0.0218
# 5 PC5 13 0.318 0.0377
# 6 PC6 11 0.561 0.041
# 7 PC7 9 0.754 0.0633
# 8 PC8 7 0.804 0.232
# 9 PC9 5 0.934 0.944
Biplots
The well-known AMMI1 and AMMI2 biplots can be created with
plot_scores().
Note that since
performs_ammi
allows analyzing multiple variables at once, e.g.,
resp = c(v1, v2, ...), the output model is a list, in this case with
two elements (GY and HM). By default, the biplots are created for the
first variable of the model. To choose another variable use the argument
var (e.g., var = "HM").
a <- plot_scores(model)
b <- plot_scores(model,
type = 2, # AMMI 2 biplot
polygon = TRUE, # show a polygon
highlight = c("G4", "G5", "G6"), #highlight genotypes
col.alpha.env = 0.5, # alpha for environments
col.alpha.gen = 0, # remove the other genotypes
col.env = "gray", # color for environment point
col.segm.env = "gray", # color for environment segment
plot_theme = theme_metan_minimal()) # theme
arrange_ggplot(a, b, tag_levels = "a")

GGE model
The GGE model is fitted with the function
gge(). For
more details, see the complete
vignette.
model <- gge(data_ge, ENV, GEN, GY)
model2 <- gge(data_ge, ENV, GEN, GY, svp = "genotype")
model3 <- gge(data_ge, ENV, GEN, GY, svp = "symmetrical")
a <- plot(model)
b <- plot(model2, type = 8)
c <- plot(model2,
type = 2,
col.gen = "black",
col.env = "gray70",
axis.expand = 1.5,
plot_theme = theme_metan_minimal())
arrange_ggplot(a, b, c, tag_levels = "a")

BLUP model
Linear-mixed effect models to predict the response variable in METs are
fitted using the function
gamem_met().
Here we will obtain the predicted means for genotypes in the variables
GY and HM. For more details, see the complete
vignette.
model2 <-
gamem_met(data_ge,
env = ENV,
gen = GEN,
rep = REP,
resp = everything())
# Evaluating trait GY |====================== | 50% 00:00:01 Evaluating trait HM |============================================| 100% 00:00:02
# Method: REML/BLUP
# Random effects: GEN, GEN:ENV
# Fixed effects: ENV, REP(ENV)
# Denominador DF: Satterthwaite's method
# ---------------------------------------------------------------------------
# P-values for Likelihood Ratio Test of the analyzed traits
# ---------------------------------------------------------------------------
# model GY HM
# COMPLETE NA NA
# GEN 1.11e-05 5.07e-03
# GEN:ENV 2.15e-11 2.27e-15
# ---------------------------------------------------------------------------
# All variables with significant (p < 0.05) genotype-vs-environment interaction
# Get the variance components
get_model_data(model2, what = "vcomp")
# Class of the model: waasb
# Variable extracted: vcomp
# # A tibble: 3 × 3
# Group GY HM
# <chr> <dbl> <dbl>
# 1 GEN 0.0280 0.490
# 2 GEN:ENV 0.0567 2.19
# 3 Residual 0.0967 2.84
Plotting the BLUPs for genotypes
To produce a plot with the predicted means, use the function
plot_blup().
a <- plot_blup(model2)
b <- plot_blup(model2,
prob = 0.2,
col.shape = c("gray20", "gray80"),
invert = TRUE)
arrange_ggplot(a, b, tag_levels = "a")

Computing parametric and non-parametric stability indexes
The easiest way to compute parametric and non-parametric stability
indexes in metan is by using the function
ge_stats().
It is a wrapper function around a lot of specific functions for
stability indexes. To get the results into a “ready-to-read” file, use
get_model_data()
or its shortcut
gmd().
stats <- ge_stats(data_ge, ENV, GEN, REP, GY)
# Evaluating trait GY |============================================| 100% 00:00:08
get_model_data(stats)
# Class of the model: ge_stats
# Variable extracted: stats
# # A tibble: 10 × 44
# var GEN Y CV ACV POLAR Var Shukla Wi_g Wi_f Wi_u Ecoval
# <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
# 1 GY G1 2.60 35.2 34.1 0.0298 10.9 0.0280 84.4 89.2 81.1 1.22
# 2 GY G10 2.47 42.3 38.6 0.136 14.2 0.244 59.2 64.6 54.4 7.96
# 3 GY G2 2.74 34.0 35.2 0.0570 11.3 0.0861 82.8 95.3 75.6 3.03
# 4 GY G3 2.96 29.9 33.8 0.0216 10.1 0.0121 104. 99.7 107. 0.725
# 5 GY G4 2.64 31.4 31.0 -0.0537 8.93 0.0640 85.9 79.5 91.9 2.34
# 6 GY G5 2.54 30.6 28.8 -0.119 7.82 0.0480 82.7 82.2 82.4 1.84
# 7 GY G6 2.53 29.7 27.8 -0.147 7.34 0.0468 83.0 83.7 81.8 1.81
# 8 GY G7 2.74 27.4 28.3 -0.133 7.33 0.122 83.9 77.6 93.4 4.16
# 9 GY G8 3.00 30.4 35.1 0.0531 10.8 0.0712 98.8 90.5 107. 2.57
# 10 GY G9 2.51 42.4 39.4 0.154 14.7 0.167 68.8 68.9 70.3 5.56
# # … with 32 more variables: bij <dbl>, Sij <dbl>, R2 <dbl>, ASTAB <dbl>,
# # ASI <dbl>, ASV <dbl>, AVAMGE <dbl>, DA <dbl>, DZ <dbl>, EV <dbl>, FA <dbl>,
# # MASI <dbl>, MASV <dbl>, SIPC <dbl>, ZA <dbl>, WAAS <dbl>, WAASB <dbl>,
# # HMGV <dbl>, RPGV <dbl>, HMRPGV <dbl>, Pi_a <dbl>, Pi_f <dbl>, Pi_u <dbl>,
# # Gai <dbl>, S1 <dbl>, S2 <dbl>, S3 <dbl>, S6 <dbl>, N1 <dbl>, N2 <dbl>,
# # N3 <dbl>, N4 <dbl>
Citation
citation("metan")
Please, support this project by citing it in your publications!
Olivoto, T., and Lúcio, A.D. (2020). metan: an R package for
multi-environment trial analysis. Methods Ecol Evol. 11:783-789
doi:10.1111/2041-210X.13384
A BibTeX entry for LaTeX users is
@Article{Olivoto2020,
author = {Tiago Olivoto and Alessandro Dal'Col L{'{u}}cio},
title = {metan: an R package for multi-environment trial analysis},
journal = {Methods in Ecology and Evolution},
volume = {11},
number = {6},
pages = {783-789},
year = {2020},
doi = {10.1111/2041-210X.13384},
}
Getting help
-
If you encounter a clear bug, please file a minimal reproducible example on github
-
Suggestions and criticisms to improve the quality and usability of the package are welcome!
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.