In brunobrr/bdc: Biodiversity Data Cleaning
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
fig.path = "man/figures/README-",
out.width = "100%",
echo = TRUE,
warning = FALSE,
eval = TRUE
)
library(bdc)
# if (!requireNamespace("rnaturalearthhires", quietly = TRUE))
# remotes::install_github("ropensci/rnaturalearthhires")
Introduction
Large and heterogeneous datasets may contain thousands of records missing spatial or taxonomic information (partially or entirely) as well as several records outside a region of interest or from doubtful sources. Such lower quality data are not fit for use in many research applications without prior amendments. The 'Pre-filter' module contains a series of tests to detect, remove, and, whenever, possible, correct such erroneous or suspect records.
Installation
Check here how to install the bdc package.
Reading the database
Read the merged database created in the module Standardization and integration of different datasets of the bdc package. It is also possible to read any datasets containing the required fields to run the functions (more details here).
database <-
readr::read_csv(here::here("Output/Intermediate/00_merged_database.csv"))
database <-
readr::read_csv(
system.file("extdata/outpus_vignettes/00_merged_database.csv", package = "bdc"),
show_col_types = FALSE
)
DT::datatable(
database[1:15,], class = 'stripe', extensions = 'FixedColumns',
rownames = FALSE,
options = list(
pageLength = 5,
dom = 'Bfrtip',
scrollX = TRUE,
fixedColumns = list(leftColumns = 2)
)
)
⚠️IMPORTANT:
The results of the VALIDATION test used to flag data quality are appended in separate fields in this database and retrieved as TRUE ( ✅ ok ) or FALSE (❌check carefully ).
1 - Records missing species names
VALIDATION. Flag records missing species names.
check_pf <-
bdc_scientificName_empty(
data = database,
sci_name = "scientificName")
#> bdc_scientificName_empty:
#> Flagged 5 records.
#> One column was added to the database.
2 - Records lacking information on geographic coordinates
VALIDATION. Flag records missing partial or complete information on geographic coordinates.
check_pf <- bdc_coordinates_empty(
data = check_pf,
lat = "decimalLatitude",
lon = "decimalLongitude")
#> bdc_coordinates_empty:
#> Flagged 44 records.
#> One column was added to the database.
3 - Records with out-of-range coordinates
VALIDATION. This test flags records with out-of-range coordinates: latitude > 90 or -90; longitude >180 or -180.
check_pf <- bdc_coordinates_outOfRange(
data = check_pf,
lat = "decimalLatitude",
lon = "decimalLongitude")
#> bdc_coordinates_outOfRange:
#> Flagged 1 records.
#> One column was added to the database.
4 - Records from doubtful sources
VALIDATION. This test flags records from doubtful source. For example, records from drawings, photographs, or multimedia objects, fossil records, among others.
# Check record sources of your dataset using:
# check_pf %>%
# dplyr::group_by(basisOfRecord) %>%
# dplyr::summarise(n = dplyr::n())
check_pf <- bdc_basisOfRecords_notStandard(
data = check_pf,
basisOfRecord = "basisOfRecord",
names_to_keep = "all")
#> bdc_basisOfRecords_notStandard:
#> Flagged 0 of the following specific nature:
#> character(0)
#> One column was added to the database.
5 - Getting country names from valid coordinates
ENRICHMENT. Deriving country names for records missing country names.
check_pf <- bdc_country_from_coordinates(
data = check_pf,
lat = "decimalLatitude",
lon = "decimalLongitude",
country = "country")
#> bdc_country_from_coordinates:
#> Country names were added to 27 records.
6 - Standardizing country names and getting country code information
ENRICHMENT. Country names are standardized against a list of country names in several languages retrieved from International Organization for Standardization https://stefangabos.github.io/world_countries/.
check_pf <- bdc_country_standardized(
data = check_pf,
country = "country"
)
#> Standardizing country names
#>
#> country found: Bolivia
#> country found: Brazil
#> |++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=00s
#> |++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=00s
#> |++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=00s
#>
#> bdc_country_standardized:
#> The country names of 65 records were standardized.
#> Two columns ('country_suggested' and 'countryCode') were added to the database.
7 - Correcting latitude and longitude transposed
AMENDMENT. The mismatch between informed country and coordinates can be the result of negative or transposed coordinates. Once detected a mismatch, different coordinate transformations are made to correct the country and coordinates mismatch. Verbatim coordinates are then replaced by the rectified ones in the returned database (a database containing verbatim and corrected coordinates can be created in the "Output" folder if save_outputs = TRUE). Records near countries coastline are not tested to avoid incur in false positives.
check_pf <-
bdc_coordinates_transposed(
data = check_pf,
id = "database_id",
sci_names = "scientificName",
lat = "decimalLatitude",
lon = "decimalLongitude",
country = "country_suggested",
countryCode = "countryCode",
border_buffer = 0.2, # in decimal degrees (~22 km at the equator)
save_outputs = FALSE
)
#> Correcting latitude and longitude transposed
#>
#> 15 occurrences will be tested
#> Processing occurrences from: BR (15)
#> No latitude and longitude were transposed
8 - Records outside a region of interest
VALIDATION. Records outside one or multiple reference countries; i.e., records in other countries or at an informed distance from the coast (e.g., in the ocean). This last step avoids flagging as invalid records close to country limits (e.g., records of coast or marshland species).
check_pf <-
bdc_coordinates_country_inconsistent(
data = check_pf,
country_name = "Brazil",
country = "country_suggested",
lon = "decimalLongitude",
lat = "decimalLatitude",
dist = 0.1 # in decimal degrees (~11 km at the equator)
)
#> dist is assumed to be in decimal degrees (arc_degrees).
#> although coordinates are longitude/latitude, st_intersection assumes that they are planar
#>
#> bdc_coordinates_country_inconsistent:
#> Flagged 15 records.
#> One column was added to the database.
9 - Identifying records not geo-referenced but containing locality information
ENRICHMENT. Coordinates can be derived from a detailed description of the locality associated with records in a process called retrospective geo-referencing.
xyFromLocality <- bdc_coordinates_from_locality(
data = check_pf,
locality = "locality",
lon = "decimalLongitude",
lat = "decimalLatitude",
save_outputs = FALSE
)
#> bdc_coordinates_from_locality
#> Found 38 records missing or with invalid coordinates but with potentially useful information on locality
Report
Here we create a column named ".summary" summing up the results of all VALIDATION tests (those starting with "."). This column is FALSE when a record is flagged as FALSE in any data quality test (i.e. potentially invalid or suspect record).
check_pf <- bdc_summary_col(data = check_pf)
#> bdc_summary_col:
#> Flagged 65 records.
#> One column was added to the database.
# DT::datatable(
# check_pf, class = 'stripe', extensions = 'FixedColumns',
# rownames = FALSE,
# options = list(
# pageLength = 3,
# dom = 'Bfrtip',
# scrollX = TRUE,
# fixedColumns = list(leftColumns = 2)
# )
# )
Creating a report summarizing the results of all tests. The report can be automatically saved if save_report = TRUE.
report <-
bdc_create_report(data = check_pf,
database_id = "database_id",
workflow_step = "prefilter",
save_report = FALSE)
report
report <-
readr::read_csv(
system.file("extdata/outpus_vignettes/01_Report_Prefilter.csv",
package = "bdc"),
show_col_types = FALSE
)
DT::datatable(
report, class = 'stripe', extensions = 'FixedColumns',
rownames = FALSE,
options = list(
# pageLength = 5,
dom = 'Bfrtip',
scrollX = TRUE,
fixedColumns = list(leftColumns = 2)
)
)
Figures
Here we create figures (bar plots and maps) to make the interpretation of the results of data quality tests easier. See some examples below. Figures can be automatically saved if save_figures = TRUE.
figures <-
bdc_create_figures(data = check_pf,
database_id = "database_id",
workflow_step = "prefilter",
save_figures = FALSE)
# Check figures using
figures$.coordinates_country_inconsistent
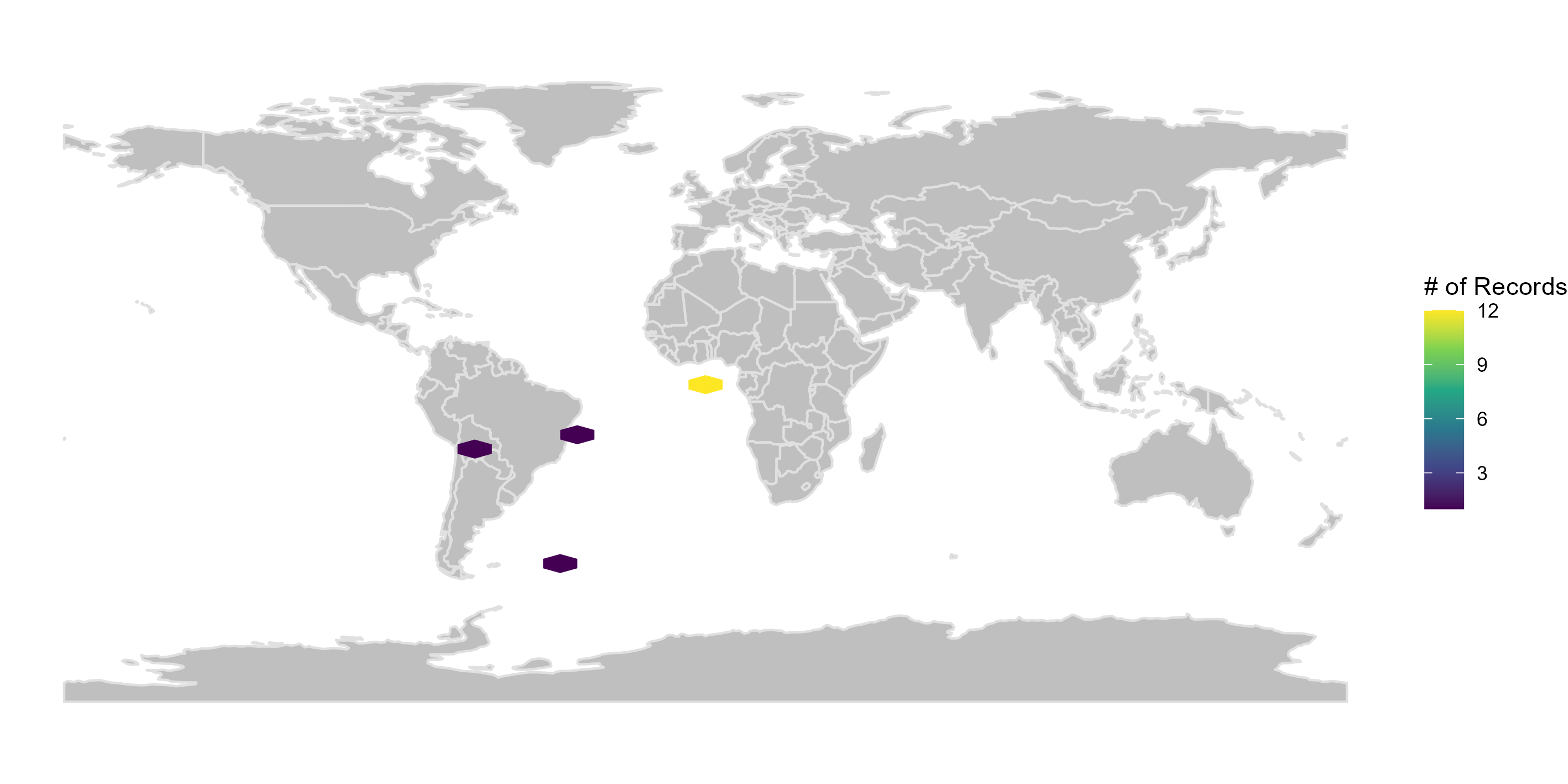
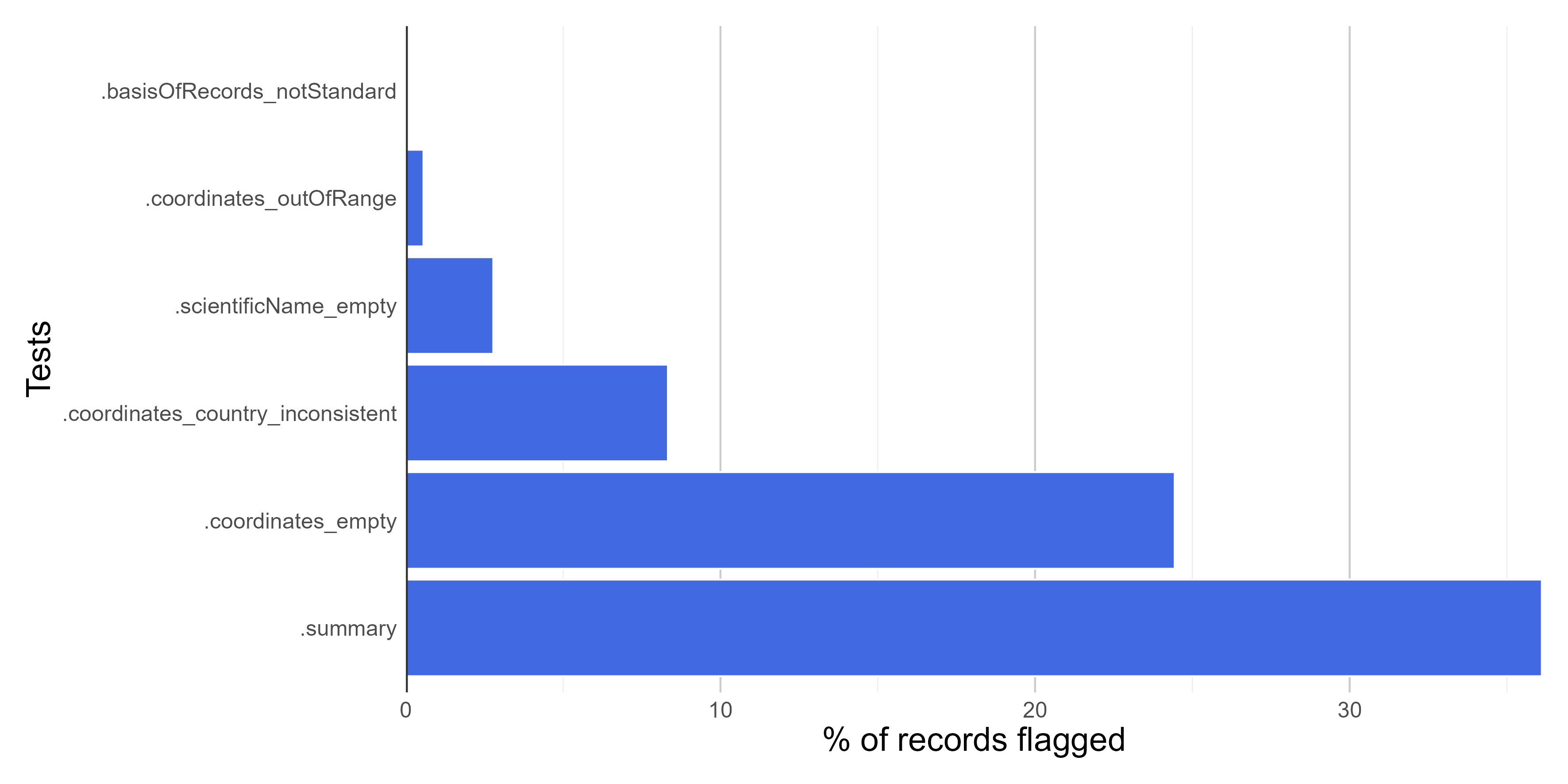
Filtering the database
We can remove records flagged as erroneous or suspect to obtain a "clean" database. Records missing names or coordinates, outside a region of interest or from distrustful sources are rarely suitable for biodiversity analyses. We can use the column .summary to filter valid records passing in all tests (i.e., flagged as "TRUE"). Next, we use the bdc_filter_out_falgs function to remove all tests' columns (that is, those starting with ".").
output <-
check_pf %>%
dplyr::filter(.summary == TRUE) %>%
bdc_filter_out_flags(data = ., col_to_remove = "all")
#> bdc_fiter_out_flags:
#> The following columns were removed from the database:
#> .scientificName_empty, .coordinates_empty, .coordinates_outOfRange, .basisOfRecords_notStandard, .coordinates_country_inconsistent, .summary
Saving the database
You can use [qs::qsave()]{.underline} instead of write_csv to save a large database in a compressed format.
# use qs::qsave() to save the database in a compressed format and then qs:qread() to load the database
output %>%
readr::write_csv(.,
here::here("Output", "Intermediate", "01_prefilter_database.csv"))
brunobrr/bdc documentation built on Nov. 21, 2024, 4:18 a.m.
knitr::opts_chunk$set( collapse = TRUE, comment = "#>", fig.path = "man/figures/README-", out.width = "100%", echo = TRUE, warning = FALSE, eval = TRUE )
library(bdc) # if (!requireNamespace("rnaturalearthhires", quietly = TRUE)) # remotes::install_github("ropensci/rnaturalearthhires")
Introduction
Large and heterogeneous datasets may contain thousands of records missing spatial or taxonomic information (partially or entirely) as well as several records outside a region of interest or from doubtful sources. Such lower quality data are not fit for use in many research applications without prior amendments. The 'Pre-filter' module contains a series of tests to detect, remove, and, whenever, possible, correct such erroneous or suspect records.
Installation
Check here how to install the bdc package.
Reading the database
Read the merged database created in the module Standardization and integration of different datasets of the bdc package. It is also possible to read any datasets containing the required fields to run the functions (more details here).
database <- readr::read_csv(here::here("Output/Intermediate/00_merged_database.csv"))
database <- readr::read_csv( system.file("extdata/outpus_vignettes/00_merged_database.csv", package = "bdc"), show_col_types = FALSE )
DT::datatable( database[1:15,], class = 'stripe', extensions = 'FixedColumns', rownames = FALSE, options = list( pageLength = 5, dom = 'Bfrtip', scrollX = TRUE, fixedColumns = list(leftColumns = 2) ) )
⚠️IMPORTANT:
The results of the VALIDATION test used to flag data quality are appended in separate fields in this database and retrieved as TRUE ( ✅ ok ) or FALSE (❌check carefully ).
1 - Records missing species names
VALIDATION. Flag records missing species names.
check_pf <- bdc_scientificName_empty( data = database, sci_name = "scientificName") #> bdc_scientificName_empty: #> Flagged 5 records. #> One column was added to the database.
2 - Records lacking information on geographic coordinates
VALIDATION. Flag records missing partial or complete information on geographic coordinates.
check_pf <- bdc_coordinates_empty( data = check_pf, lat = "decimalLatitude", lon = "decimalLongitude") #> bdc_coordinates_empty: #> Flagged 44 records. #> One column was added to the database.
3 - Records with out-of-range coordinates
VALIDATION. This test flags records with out-of-range coordinates: latitude > 90 or -90; longitude >180 or -180.
check_pf <- bdc_coordinates_outOfRange( data = check_pf, lat = "decimalLatitude", lon = "decimalLongitude") #> bdc_coordinates_outOfRange: #> Flagged 1 records. #> One column was added to the database.
4 - Records from doubtful sources
VALIDATION. This test flags records from doubtful source. For example, records from drawings, photographs, or multimedia objects, fossil records, among others.
# Check record sources of your dataset using: # check_pf %>% # dplyr::group_by(basisOfRecord) %>% # dplyr::summarise(n = dplyr::n()) check_pf <- bdc_basisOfRecords_notStandard( data = check_pf, basisOfRecord = "basisOfRecord", names_to_keep = "all") #> bdc_basisOfRecords_notStandard: #> Flagged 0 of the following specific nature: #> character(0) #> One column was added to the database.
5 - Getting country names from valid coordinates
ENRICHMENT. Deriving country names for records missing country names.
check_pf <- bdc_country_from_coordinates( data = check_pf, lat = "decimalLatitude", lon = "decimalLongitude", country = "country") #> bdc_country_from_coordinates: #> Country names were added to 27 records.
6 - Standardizing country names and getting country code information
ENRICHMENT. Country names are standardized against a list of country names in several languages retrieved from International Organization for Standardization https://stefangabos.github.io/world_countries/.
check_pf <- bdc_country_standardized( data = check_pf, country = "country" ) #> Standardizing country names #> #> country found: Bolivia #> country found: Brazil #> |++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=00s #> |++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=00s #> |++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=00s #> #> bdc_country_standardized: #> The country names of 65 records were standardized. #> Two columns ('country_suggested' and 'countryCode') were added to the database.
7 - Correcting latitude and longitude transposed
AMENDMENT. The mismatch between informed country and coordinates can be the result of negative or transposed coordinates. Once detected a mismatch, different coordinate transformations are made to correct the country and coordinates mismatch. Verbatim coordinates are then replaced by the rectified ones in the returned database (a database containing verbatim and corrected coordinates can be created in the "Output" folder if save_outputs = TRUE). Records near countries coastline are not tested to avoid incur in false positives.
check_pf <- bdc_coordinates_transposed( data = check_pf, id = "database_id", sci_names = "scientificName", lat = "decimalLatitude", lon = "decimalLongitude", country = "country_suggested", countryCode = "countryCode", border_buffer = 0.2, # in decimal degrees (~22 km at the equator) save_outputs = FALSE ) #> Correcting latitude and longitude transposed #> #> 15 occurrences will be tested #> Processing occurrences from: BR (15) #> No latitude and longitude were transposed
8 - Records outside a region of interest
VALIDATION. Records outside one or multiple reference countries; i.e., records in other countries or at an informed distance from the coast (e.g., in the ocean). This last step avoids flagging as invalid records close to country limits (e.g., records of coast or marshland species).
check_pf <- bdc_coordinates_country_inconsistent( data = check_pf, country_name = "Brazil", country = "country_suggested", lon = "decimalLongitude", lat = "decimalLatitude", dist = 0.1 # in decimal degrees (~11 km at the equator) ) #> dist is assumed to be in decimal degrees (arc_degrees). #> although coordinates are longitude/latitude, st_intersection assumes that they are planar #> #> bdc_coordinates_country_inconsistent: #> Flagged 15 records. #> One column was added to the database.
9 - Identifying records not geo-referenced but containing locality information
ENRICHMENT. Coordinates can be derived from a detailed description of the locality associated with records in a process called retrospective geo-referencing.
xyFromLocality <- bdc_coordinates_from_locality( data = check_pf, locality = "locality", lon = "decimalLongitude", lat = "decimalLatitude", save_outputs = FALSE ) #> bdc_coordinates_from_locality #> Found 38 records missing or with invalid coordinates but with potentially useful information on locality
Report
Here we create a column named ".summary" summing up the results of all VALIDATION tests (those starting with "."). This column is FALSE when a record is flagged as FALSE in any data quality test (i.e. potentially invalid or suspect record).
check_pf <- bdc_summary_col(data = check_pf) #> bdc_summary_col: #> Flagged 65 records. #> One column was added to the database.
# DT::datatable( # check_pf, class = 'stripe', extensions = 'FixedColumns', # rownames = FALSE, # options = list( # pageLength = 3, # dom = 'Bfrtip', # scrollX = TRUE, # fixedColumns = list(leftColumns = 2) # ) # )
Creating a report summarizing the results of all tests. The report can be automatically saved if save_report = TRUE.
report <- bdc_create_report(data = check_pf, database_id = "database_id", workflow_step = "prefilter", save_report = FALSE) report
report <- readr::read_csv( system.file("extdata/outpus_vignettes/01_Report_Prefilter.csv", package = "bdc"), show_col_types = FALSE )
DT::datatable( report, class = 'stripe', extensions = 'FixedColumns', rownames = FALSE, options = list( # pageLength = 5, dom = 'Bfrtip', scrollX = TRUE, fixedColumns = list(leftColumns = 2) ) )
Figures
Here we create figures (bar plots and maps) to make the interpretation of the results of data quality tests easier. See some examples below. Figures can be automatically saved if save_figures = TRUE.
figures <- bdc_create_figures(data = check_pf, database_id = "database_id", workflow_step = "prefilter", save_figures = FALSE) # Check figures using figures$.coordinates_country_inconsistent


Filtering the database
We can remove records flagged as erroneous or suspect to obtain a "clean" database. Records missing names or coordinates, outside a region of interest or from distrustful sources are rarely suitable for biodiversity analyses. We can use the column .summary to filter valid records passing in all tests (i.e., flagged as "TRUE"). Next, we use the bdc_filter_out_falgs function to remove all tests' columns (that is, those starting with ".").
output <- check_pf %>% dplyr::filter(.summary == TRUE) %>% bdc_filter_out_flags(data = ., col_to_remove = "all") #> bdc_fiter_out_flags: #> The following columns were removed from the database: #> .scientificName_empty, .coordinates_empty, .coordinates_outOfRange, .basisOfRecords_notStandard, .coordinates_country_inconsistent, .summary
Saving the database
You can use [qs::qsave()]{.underline} instead of write_csv to save a large database in a compressed format.
# use qs::qsave() to save the database in a compressed format and then qs:qread() to load the database output %>% readr::write_csv(., here::here("Output", "Intermediate", "01_prefilter_database.csv"))
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
