README.md
In natverse/insectbrainr: R client utilities for interacting with the InsectBrainDB.org
The Insect Brain Database
The goal of insectbrainr is to provide R client utilities for
interacting with the Insect Brain
Database. Using this R package in
concert with the natverse
ecosystem of neuroanatomy tools is highly recommended. The
InsectBrainDB.org is primarily curated
by Stanley Heinze. Learn
more about the project here.
Installation
Firstly, you will need R, R Studio and X Quartz as well as nat and its
dependencies. For detailed installation instructions for all this, see
here. It
should not take too long at all. Then:
# install
if (!require("remotes")) install.packages("remotes")
remotes::install_github("natverse/insectbrainr")
# use
library(insectbrainr)
Done!
Key Functions
Now we can have a look at what is available, here are some of the key
functions. Their help details examples of their use. You can summon the
help in RStudio using ? followed by the function name.
# And how can I read neurons from the insectbrainDB?
?insectbrainr_read_neurons()
# Get 3D neuropil-subdivided brain models for those brainspaces
?insectbraindb_read_brain # Get 3D neuropil-subdivided brain models for those brainspaces
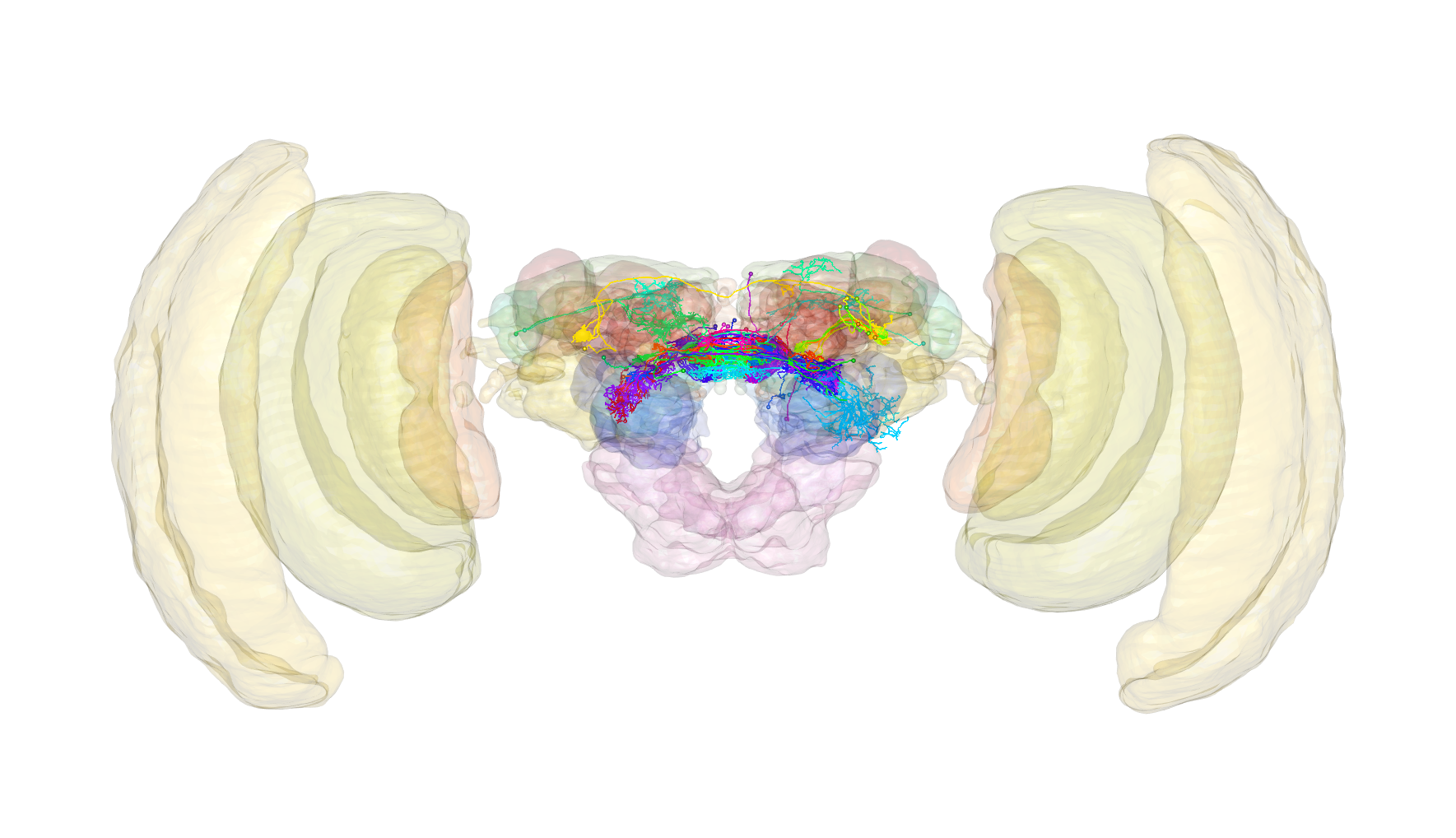
Example
Let’s also have a look at an example pulling neurons and brain meshes
from insectbraindb.org. Here we shall
take a look at neurons from the brain of the Monarch butterfly that have
been registered to a template brain. Excitingly, we can also visualise
this template brain.
## What neurons does the insectbraindb.org host?
available.neurons = insectbraindb_neuron_info()
## Let's just download all of the neurons in the database to play with,
## there are not very many:
nrow(available.neurons)
## First, we call the read neurons function, with ids set to NULL
insect.neurons = insectbraindb_read_neurons(ids = NULL)
## Hmm, let's see how many neurons we have perspecies
table(insect.neurons[,"common_name"])
## So, it seem the Monarch Butterfly is the clear winner there,
## maybe let's just have those
butterfly.neurons = subset(insect.neurons, common_name == "Monarch Butterfly")
## And let's plot them
nat::nopen3d(userMatrix = structure(c(0.999986588954926, -0.00360279157757759,
-0.00371213257312775, 0, -0.00464127957820892, -0.941770493984222,
-0.336223870515823, 0, -0.00228461623191833, 0.336236596107483,
-0.941774606704712, 0, 0, 0, 0, 1), .Dim = c(4L, 4L)), zoom = 0.600000023841858,
windowRect = c(1460L, 65L, 3229L, 1083L))
plot3d(butterfly.neurons, lwd = 2, soma = 5)
## Cool! But maybe we also want to see it's template brain?
## Let's check if they have it
available.brains = insectbraindb_species_info()
available.brains
## Great, they do, let's get it
butterfly.brain = insectbraindb_read_brain(species = "Danaus plexippus")
## And plot in a translucent manner
plot3d(butterfly.brain, alpha = 0.1)
## Oop, that's a lot of neuropils.
## Let's go for only a subset. What's available?
butterfly.brain$RegionList
butterfly.brain$neuropil_full_names
## There lateral horn (LH) and the antennal lobe (AL) are my favourites.
## Let's plot those
clear3d()
plot3d(subset(butterfly.brain, "LH|AL"), alpha = 0.5)
plot3d(butterfly.neurons, lwd = 2, soma = 5)
### Ffff, doesn't look like we have any neurons in my favourite neuropils :(
Acknowledging the data and tools
The insectbraindb.org has a terms of
use, which provides guidance on
how best to credit data from these repositories. Most neurons have an
associated publication that you can find on the repository websites.
This package was created by Alexander Shakeel Bates, while in the group
of Dr. Gregory
Jefferis. You can cite
this package as:
citation(package = "insectbrainr")
Bates AS (2019). insectbrainr: R client utilities for interacting
with the InsectBrainDB.org. R package version 0.1.0.
https://github.com/natverse/insectbrainr
Acknowledgements
The insectbraindb.org is primarily
curated by Dr. Stanley
Heinze, and was built by
Kevin Tedore, and has several significant
supporters, including the ERC.
natverse/insectbrainr documentation built on April 24, 2023, 6:06 a.m.
The Insect Brain Database
The goal of insectbrainr is to provide R client utilities for interacting with the Insect Brain Database. Using this R package in concert with the natverse ecosystem of neuroanatomy tools is highly recommended. The InsectBrainDB.org is primarily curated by Stanley Heinze. Learn more about the project here.
Installation
Firstly, you will need R, R Studio and X Quartz as well as nat and its dependencies. For detailed installation instructions for all this, see here. It should not take too long at all. Then:
# install
if (!require("remotes")) install.packages("remotes")
remotes::install_github("natverse/insectbrainr")
# use
library(insectbrainr)
Done!
Key Functions
Now we can have a look at what is available, here are some of the key
functions. Their help details examples of their use. You can summon the
help in RStudio using ? followed by the function name.
# And how can I read neurons from the insectbrainDB?
?insectbrainr_read_neurons()
# Get 3D neuropil-subdivided brain models for those brainspaces
?insectbraindb_read_brain # Get 3D neuropil-subdivided brain models for those brainspaces
Example
Let’s also have a look at an example pulling neurons and brain meshes from insectbraindb.org. Here we shall take a look at neurons from the brain of the Monarch butterfly that have been registered to a template brain. Excitingly, we can also visualise this template brain.
## What neurons does the insectbraindb.org host?
available.neurons = insectbraindb_neuron_info()
## Let's just download all of the neurons in the database to play with,
## there are not very many:
nrow(available.neurons)
## First, we call the read neurons function, with ids set to NULL
insect.neurons = insectbraindb_read_neurons(ids = NULL)
## Hmm, let's see how many neurons we have perspecies
table(insect.neurons[,"common_name"])
## So, it seem the Monarch Butterfly is the clear winner there,
## maybe let's just have those
butterfly.neurons = subset(insect.neurons, common_name == "Monarch Butterfly")
## And let's plot them
nat::nopen3d(userMatrix = structure(c(0.999986588954926, -0.00360279157757759,
-0.00371213257312775, 0, -0.00464127957820892, -0.941770493984222,
-0.336223870515823, 0, -0.00228461623191833, 0.336236596107483,
-0.941774606704712, 0, 0, 0, 0, 1), .Dim = c(4L, 4L)), zoom = 0.600000023841858,
windowRect = c(1460L, 65L, 3229L, 1083L))
plot3d(butterfly.neurons, lwd = 2, soma = 5)
## Cool! But maybe we also want to see it's template brain?
## Let's check if they have it
available.brains = insectbraindb_species_info()
available.brains
## Great, they do, let's get it
butterfly.brain = insectbraindb_read_brain(species = "Danaus plexippus")
## And plot in a translucent manner
plot3d(butterfly.brain, alpha = 0.1)
## Oop, that's a lot of neuropils.
## Let's go for only a subset. What's available?
butterfly.brain$RegionList
butterfly.brain$neuropil_full_names
## There lateral horn (LH) and the antennal lobe (AL) are my favourites.
## Let's plot those
clear3d()
plot3d(subset(butterfly.brain, "LH|AL"), alpha = 0.5)
plot3d(butterfly.neurons, lwd = 2, soma = 5)
### Ffff, doesn't look like we have any neurons in my favourite neuropils :(

Acknowledging the data and tools
The insectbraindb.org has a terms of use, which provides guidance on how best to credit data from these repositories. Most neurons have an associated publication that you can find on the repository websites.
This package was created by Alexander Shakeel Bates, while in the group of Dr. Gregory Jefferis. You can cite this package as:
citation(package = "insectbrainr")
Bates AS (2019). insectbrainr: R client utilities for interacting with the InsectBrainDB.org. R package version 0.1.0. https://github.com/natverse/insectbrainr
Acknowledgements
The insectbraindb.org is primarily curated by Dr. Stanley Heinze, and was built by Kevin Tedore, and has several significant supporters, including the ERC.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.


