README.md
In tejaslodaya/troop: Groupby and Apply Function to a data.table Using Parallel Processing
Getting Started
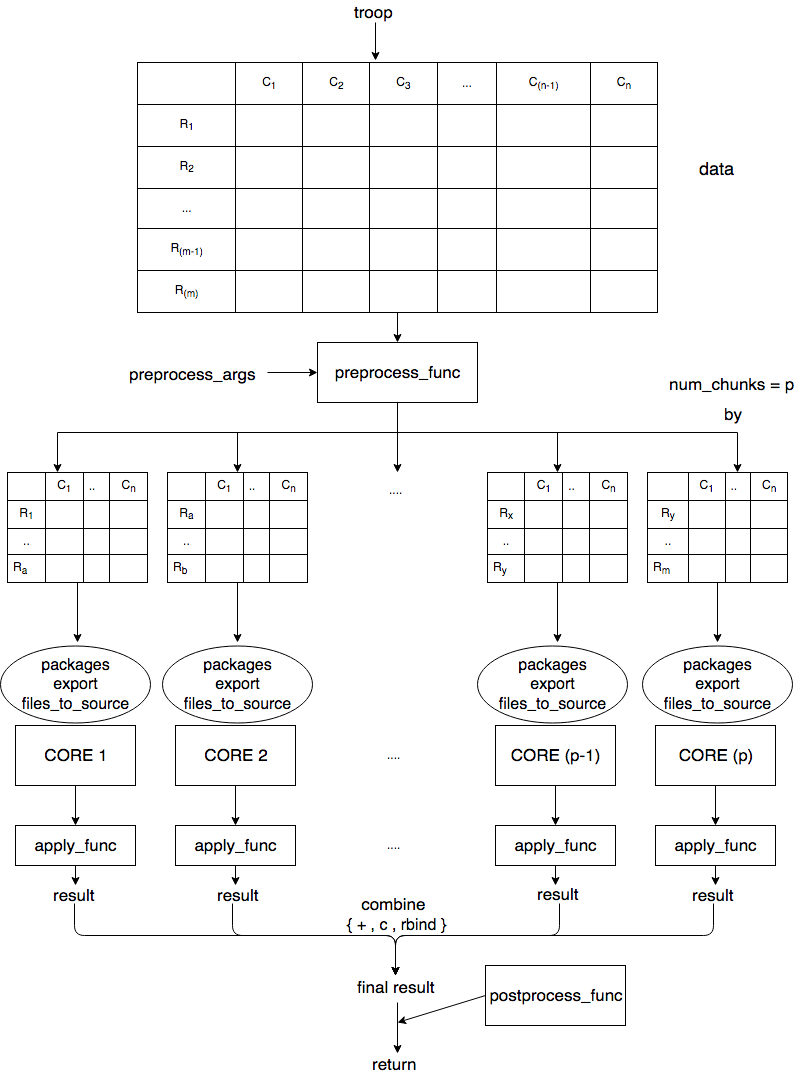
Logic behind this package is explained in the image below
Features
- Identify repeated patterns in R code and exploit to greatly improve efficiency, both within a multi-core machine and for communication across machines.
- Replace dynamic cache and server structures with static pre-serialized structures
- Pre-fetch information and determine which data should be cached at each thread to avoid contention and slow access to memory banks attached to sockets.
troop follows a data-parallel approach rather than a model-parallel approach where the training data is divided among worker threads that execute in parallel, each performing the work associated with their shard and communicating updates after completing task over that shard. This is also called SIMD approach (Single Instruction, Multiple Data), using the doParallel package of R. SOCK clusters are created and a chunk of data runs on each cluster.

Parameters
- Input data of type data.table (
data)
- Character vector giving columns to group by (
by)
- Function to be run in parallel (
apply_func)
- Function that will be run before apply_func. Use it to open file/db handles(
preprocess_func)
- Function that will be run after apply_func. useful to close file/db handles (
postprocess_func)
- Number of chunks to divide the data into. defaults to number of logical cores available (
num_chunks)
- A list of args to be passed to preprocess_func (
preprocess_args)
- A list of args to be passed to postprocess_func (
postprocess_args)
- Character vector of package names to be exported on each core. Each package used by
apply_func should be included (packages)
- Character vector of variable names to be exported on each core. Each variable name to be accessed inside
apply_func should be exported (export)
- The way results should be combined. Accepts: c, +, rbind. Defaults to c (
combine)
- Character vector of file names to be sourced on each core. The user should have permission to read the file (
files_to_source)
Prerequisites
R (>= 3.3.2)
Installation
troop can directly be installed from github
install.packages("devtools")
devtools::install_github("tejaslodaya/troop")
Thats it! Now you can use the package on your machine
Example
Barebone example
library(data.table)
dt <- data.table(fread('sample.csv'))
resR <- troop::troop(dt, by = c('column1','column2'), apply_func = nrow)
Complex example
library(data.table)
dt <- data.table(fread('sample.csv'))
var <- 10
foo <- function(data_chunk){
# some complex operation
resR <- summary(data_chunk)
return (resR)
}
#source file on each core
result <- troop::troop(dt, by = c('column1','column2'), apply_func = foo, files_to_source = c('somefile.R','anotherfile.R'))
#using packages and exporting variables
result <- troop::troop(dt, by = c('column1','column2'), apply_func = foo, num_chunks = 10, packages = c('RODBC','xgboost'), export = c('var'), combine = 'c')
NOTE
- Complete documentation of the method can be found by executing
?troop::troop in R console
- All variables which are used in apply_func method have to be included in export parameter
- All packages which are used in apply_func method have to be included in packages parameter
- If num_chunks passed is less than total number of combinations, in that case each core will execute more than one combination sequentially
TODO
- Straggler Mitigation - Each time synchronization is required, any one slowed worker thread can cause significant unproductive wait time for the others. Troop should temporarily offload a portion of its work to workers that are currently faster, helping the slowed worker catch up.
Built With
- draw.io - website used to create image
- devtools - making developer's life easy
- roxygen - generate documentation
Contributing
Please read CONTRIBUTING.md for details on our code of conduct, and the process for submitting pull requests to us.
Authors
- Tejas Lodaya - tejaslodaya.github.io
License
This project is licensed under the MIT License
References
- http://stat.ethz.ch/R-manual/R-devel/library/parallel/doc/parallel.pdf
- http://r.adu.org.za/web/packages/foreach/vignettes/foreach.pdf
- https://cran.r-project.org/web/packages/doParallel/vignettes/gettingstartedParallel.pdf
- http://michaeljkoontz.weebly.com/uploads/1/9/9/4/19940979/parallel.pdf
- https://cran.r-project.org/web/packages/iterators/vignettes/writing.pdf
tejaslodaya/troop documentation built on March 6, 2023, 11:44 p.m.
Getting Started
Logic behind this package is explained in the image below

Features
- Identify repeated patterns in R code and exploit to greatly improve efficiency, both within a multi-core machine and for communication across machines.
- Replace dynamic cache and server structures with static pre-serialized structures
- Pre-fetch information and determine which data should be cached at each thread to avoid contention and slow access to memory banks attached to sockets.
troop follows a data-parallel approach rather than a model-parallel approach where the training data is divided among worker threads that execute in parallel, each performing the work associated with their shard and communicating updates after completing task over that shard. This is also called SIMD approach (Single Instruction, Multiple Data), using the doParallel package of R. SOCK clusters are created and a chunk of data runs on each cluster.
Parameters
- Input data of type data.table (
data) - Character vector giving columns to group by (
by) - Function to be run in parallel (
apply_func) - Function that will be run before apply_func. Use it to open file/db handles(
preprocess_func) - Function that will be run after apply_func. useful to close file/db handles (
postprocess_func) - Number of chunks to divide the data into. defaults to number of logical cores available (
num_chunks) - A list of args to be passed to preprocess_func (
preprocess_args) - A list of args to be passed to postprocess_func (
postprocess_args) - Character vector of package names to be exported on each core. Each package used by
apply_funcshould be included (packages) - Character vector of variable names to be exported on each core. Each variable name to be accessed inside
apply_funcshould be exported (export) - The way results should be combined. Accepts: c, +, rbind. Defaults to c (
combine) - Character vector of file names to be sourced on each core. The user should have permission to read the file (
files_to_source)
Prerequisites
R (>= 3.3.2)
Installation
troop can directly be installed from github
install.packages("devtools")
devtools::install_github("tejaslodaya/troop")
Thats it! Now you can use the package on your machine
Example
Barebone example
library(data.table)
dt <- data.table(fread('sample.csv'))
resR <- troop::troop(dt, by = c('column1','column2'), apply_func = nrow)
Complex example
library(data.table)
dt <- data.table(fread('sample.csv'))
var <- 10
foo <- function(data_chunk){
# some complex operation
resR <- summary(data_chunk)
return (resR)
}
#source file on each core
result <- troop::troop(dt, by = c('column1','column2'), apply_func = foo, files_to_source = c('somefile.R','anotherfile.R'))
#using packages and exporting variables
result <- troop::troop(dt, by = c('column1','column2'), apply_func = foo, num_chunks = 10, packages = c('RODBC','xgboost'), export = c('var'), combine = 'c')
NOTE
- Complete documentation of the method can be found by executing
?troop::troopin R console - All variables which are used in apply_func method have to be included in export parameter
- All packages which are used in apply_func method have to be included in packages parameter
- If num_chunks passed is less than total number of combinations, in that case each core will execute more than one combination sequentially
TODO
- Straggler Mitigation - Each time synchronization is required, any one slowed worker thread can cause significant unproductive wait time for the others. Troop should temporarily offload a portion of its work to workers that are currently faster, helping the slowed worker catch up.
Built With
- draw.io - website used to create image
- devtools - making developer's life easy
- roxygen - generate documentation
Contributing
Please read CONTRIBUTING.md for details on our code of conduct, and the process for submitting pull requests to us.
Authors
- Tejas Lodaya - tejaslodaya.github.io
License
This project is licensed under the MIT License
References
- http://stat.ethz.ch/R-manual/R-devel/library/parallel/doc/parallel.pdf
- http://r.adu.org.za/web/packages/foreach/vignettes/foreach.pdf
- https://cran.r-project.org/web/packages/doParallel/vignettes/gettingstartedParallel.pdf
- http://michaeljkoontz.weebly.com/uploads/1/9/9/4/19940979/parallel.pdf
- https://cran.r-project.org/web/packages/iterators/vignettes/writing.pdf
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
