README.md
In vandomed/pooling: Fit Poolwise Regression Models
Fit Poolwise Regression Models
Dane Van Domelen vandomed@gmail.com
2020-02-10
Background
Pooling refers to the process of measuring a biomarker in combined
samples (e.g. blood, saliva) from multiple subjects rather than for each
subject separately. The main benefit is drastically better statistical
efficiency in certain scenarios, particularly when the per-assay cost is
high relative to other per-subject costs.
Although not used very frequently, one high-profile application of
pooling was the National Health and Nutrition Examination Survey
(NHANES), where 8-specimen pools were used in 2005-2006 to reduce the
number of assays by nearly 90% (saving $2.78 million) (Caudill 2012).
The purpose of the pooling package is to provide tools for designing
pooling studies (e.g. choosing the pool size and number of assays) and
for analyzing data from them.
Install
You can install pooling from CRAN:
install.packages("pooling")
Or from GitHub:
library("devtools")
install_github("vandomed/pooling")
Designing a pooling study
Pooling works great in the two-sample t-test scenario, because it
reduces the variance of each observation from
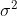 to
to
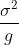 , where
, where
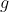 is the pool size.
is the pool size.
The poolcost_t function lets you plot total study costs vs. pool
size. To illustrate, suppose you want 80% power to detect a difference
in group means of 0.25, and the biomarker has variance 1. If it costs
$100 per assay and $10 in other per-subject recruitment costs, this is
what it looks like:
p <- poolcost_t(d = 0.25, sigsq = 1, assay_cost = 100, other_costs = 10)
p

References
Caudill, Samuel P. 2012. “Use of Pooled Samples from the National Health
and Nutrition Examination Survey.” *Statistics in Medicine* 31 (27):
3269–77.
vandomed/pooling documentation built on Feb. 22, 2020, 8:58 p.m.
Fit Poolwise Regression Models
Dane Van Domelen vandomed@gmail.com 2020-02-10
Background
Pooling refers to the process of measuring a biomarker in combined samples (e.g. blood, saliva) from multiple subjects rather than for each subject separately. The main benefit is drastically better statistical efficiency in certain scenarios, particularly when the per-assay cost is high relative to other per-subject costs.
Although not used very frequently, one high-profile application of pooling was the National Health and Nutrition Examination Survey (NHANES), where 8-specimen pools were used in 2005-2006 to reduce the number of assays by nearly 90% (saving $2.78 million) (Caudill 2012).
The purpose of the pooling package is to provide tools for designing pooling studies (e.g. choosing the pool size and number of assays) and for analyzing data from them.
Install
You can install pooling from CRAN:
install.packages("pooling")
Or from GitHub:
library("devtools")
install_github("vandomed/pooling")
Designing a pooling study
Pooling works great in the two-sample t-test scenario, because it
reduces the variance of each observation from
to
, where
is the pool size.
The poolcost_t function lets you plot total study costs vs. pool size. To illustrate, suppose you want 80% power to detect a difference in group means of 0.25, and the biomarker has variance 1. If it costs $100 per assay and $10 in other per-subject recruitment costs, this is what it looks like:
p <- poolcost_t(d = 0.25, sigsq = 1, assay_cost = 100, other_costs = 10)
p

References
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
