README.md
In cofad: Contrast Analyses for Factorial Designs
Cofad User Guide

To cite cofad in publications use:
Titz J. & Burkhardt M. (2021). cofad: An R package and shiny app for
contrast analysis. Journal of Open Source Software, 6(67), 3822,
https://doi.org/10.21105/joss.03822
A BibTeX entry for LaTeX users is
@article{titz2021,
doi = {10.21105/joss.03822},
url = {https://doi.org/10.21105/joss.03822},
year = {2021},
publisher = {The Open Journal},
volume = {6},
number = {67},
pages = {3822},
author = {Johannes Titz and Markus Burkhardt},
title = {cofad: An R package and shiny app for contrast analysis},
journal = {Journal of Open Source Software} }
Introduction
Cofad is an R package for conducting COntrast analysis in FActorial
Designs like ANOVAs. If contrast analysis was to win a price it would be
the one for the most underestimated, underused statistical technique.
This is unfortunate because in every case a contrast analysis is at
least as good as an ANOVA, but in most cases it is better. Contrast
analysis gets rid off the unspecific omnibus-hypothesis there are
differences somewhere and replaces it with a very specific numerical
hypothesis. Furthermore, contrast analysis focuses on effects instead of
significance. This is expressed doubly: First, there are three different
effect sizes for contrast analysis:
 ,
,
 and
and
 .
Second, the effect size refers not to the data but to the tested
hypothesis. The larger the effect, the more this speaks for the
hypothesis. One can even compare different hypotheses against each other
(experimentum crucis!) by looking at the effect size for each
hypothesis.
.
Second, the effect size refers not to the data but to the tested
hypothesis. The larger the effect, the more this speaks for the
hypothesis. One can even compare different hypotheses against each other
(experimentum crucis!) by looking at the effect size for each
hypothesis.
Sounds interesting? Then check out some introductory literature such as
Furr (2004), Rosenthal & Rosnow (1985), Rosenthal, Rosnow, & Rubin
(2000), or, for the German-speaking audience, Sedlmeier & Renkewitz
(2018). Contrast analysis is fairly easy to understand if you know what
an ANOVA and a correlation is. In this vignette we assume you are
familiar with the basics of contrast analysis and want to apply it to a
specific data set. First we show how to install cofad and use the
graphical user interface. Then we demonstrate some exemplary analyses
for between, within and mixed designs in R.
Installation
Cofad has two components, the plain R package and a shiny-app that
offers an intuitive graphical user interface.
If you just want to use the cofad-app, you do not need to install it.
Just go to https://cofad.titz.science and use it there. An example
data file is loaded when you add /example after the url.
If you prefer the command line interface or want to use the cofad-app
locally, install it from CRAN:
install.packages("cofad")
Alternatively, you can also install the development version from github
(you need the package devtools for this):
# install.packages("devtools") # uncomment if you do not have devtools installed
devtools::install_github("johannes-titz/cofad")
Now you can load cofad and use it in your R scripts.
You can also run the app:
cofad::run_app()
Using cofad
Before we start: Your data has to be in the long-format (also referred
to as narrow or tidy)! If you do not know what this means, please check
the short description of the Wikipedia-article:
https://en.wikipedia.org/wiki/Wide_and_narrow_data
Graphical-User-Interface
The graphical-user-interface is self-explanatory. Just load your data
and drag the variables to the correct position. At the moment you can
only read .sav (SPSS) and .csv files.
As an example go to https://cofad.titz.science/example which will load
a data set from Rosenthal et al. (2000) (Table 5.3). The cognitive
ability of nine children belonging to different age groups (between) was
measured four times (within).
There are two hypotheses:
- cognitive ability linearly increases over time (within)
(
 )
)
- cognitive ability linearly increase over age groups (between)
(
 )
)
Now drag the variables to the correct position and set the lambdas
accordingly:
 cofad GUI
cofad GUI
The result should look like this:
 cofad GUI
cofad GUI
A mixed design is ideal for testing out the cofad-app. You can now
construct a separate within-model by removing the between variable
“age”. Then you can construct a separate between-model by removing
“time” from within and dragging “age” back into the between panel.
The graphical user interface will suffice for most users, but some will
prefer to use the scripting capabilities of R. In the next sections we
will look at several script examples for different designs.
Between-Subjects Designs
Let us first load the package:
library(cofad)
Now we need some data and hypotheses. We can simply take the data from
Furr (2004), where we have different empathy ratings of students from
different majors. This data set is available in the cofad package:
data("furr_p4")
furr_p4
#> empathy major
#> 1 51 psychology
#> 2 56 psychology
#> 3 61 psychology
#> 4 58 psychology
#> 5 54 psychology
#> 6 62 education
#> 7 67 education
#> 8 57 education
#> 9 65 education
#> 10 59 education
#> 11 50 business
#> 12 49 business
#> 13 47 business
#> 14 45 business
#> 15 44 business
#> 16 50 chemistry
#> 17 45 chemistry
#> 18 40 chemistry
#> 19 49 chemistry
#> 20 41 chemistry
Furr states three hypotheses:
- Contrast A: Psychology majors have higher empathy scores than
Education majors
(
 ).
).
- Contrast B: Business majors have higher empathy scores than Chemistry
majors
(
 ).
).
- Contrast C: On average, Psychology and Education majors have higher
empathy scores than Business and Chemistry majors
(
 ).
).
These hypotheses are only mean comparisons, but this is a good way to
start. Let’s use cofad to conduct the contrast analysis:
ca <- calc_contrast(dv = empathy, between = major,
lambda_between = c("psychology" = 1, "education" = -1,
"business" = 0, "chemistry" = 0),
data = furr_p4)
ca
#>
#> We ran a contrast analysis for the following between contrasts: business = 0; chemistry = 0; education = -1; psychology = 1. This resulted in statistics of F(1,16) = 6.154; p = 0.02461 and an effect magnitude of r_effectsize = -0.276. Attention: Contrast fits in the opposite direction!
The print method shows some basic information that can be directly used
in a publication. With the summary method some more details are shown:
summary(ca)
#> Contrast Analysis Between
#>
#> Your contrast estimate is negative. This means that your data does not reflect the expected direction of your hypothesis specified by the contrast weights (lambdas).
#>
#> $Lambdas
#> business chemistry education psychology
#> 0 0 -1 1
#>
#> $tTable
#> L df t p(t≥-2.481)①
#> -6 16 -2.481 0.988
#>
#> ①The p-value refers to a one-tailed test.
#>
#> $FTable
#> SS df MS F p
#> contrast 90 1 90.000 6.154 0.0246
#> within 234 16 14.625
#> total 1179 19
#>
#> $Effects
#> effects
#> r_effectsize -0.276
#> r_contrast -0.527
#> r_alerting -0.309
From this table,
 is probably the most useful statistic. It is just the correlation
between the lambdas and the dependent variable, which can also be
calculated by hand:
is probably the most useful statistic. It is just the correlation
between the lambdas and the dependent variable, which can also be
calculated by hand:
lambdas <- rep(c(1, -1, 0, 0), each = 5)
cor(furr_p4$empathy, lambdas)
#> [1] -0.2762895
As you can see, the effect is negative and cofad also warns the user
that the contrast fits in the opposite direction. This is a big failure
for the hypothesis and indicates substantial problems in theorizing.
The other two hypotheses can be tested accordingly:
ca <- calc_contrast(dv = empathy, between = major,
lambda_between = c("psychology" = 0, "education" = 0,
"business" = 1, "chemistry" = -1),
data = furr_p4)
ca
#>
#> We ran a contrast analysis for the following between contrasts: business = 1; chemistry = -1; education = 0; psychology = 0. This resulted in statistics of F(1,16) = 0.684; p = 0.4205 and an effect magnitude of r_effectsize = 0.092.
ca <- calc_contrast(dv = empathy, between = major,
lambda_between = c("psychology" = 1, "education" = 1,
"business" = -1, "chemistry" = -1),
data = furr_p4)
ca
#>
#> We ran a contrast analysis for the following between contrasts: business = -1; chemistry = -1; education = 1; psychology = 1. This resulted in statistics of F(1,16) = 57.778; p = 0.00000107 and an effect magnitude of r_effectsize = 0.847.
When you compare the numbers to the ones presented in Furr (2004), you
will find the same result, except that Furr (2004) uses t-values and the
p-values are halved. This is because in contrast analysis you can always
test one-sided. The assumption is that your lambdas covariate positively
with the mean values, not that they either covariate positively or
negatively. Thus, you can always halve the p-value from the F-Test.
Now, imagine we have a more fun hypothesis and not just mean
differences. From an elaborate theory we could derive that the means
should be 73, 61, 51 and 38. We can test this with cofad directly
because cofad will center the lambdas (the mean of the lambdas has to be
0):
ca <- calc_contrast(dv = empathy, between = major,
lambda_between = c("psychology" = 73, "education" = 61,
"business" = 51, "chemistry" = 38),
data = furr_p4)
#> lambdas are centered and rounded to 3 digits
ca
#>
#> We ran a contrast analysis for the following between contrasts: business = -4.75; chemistry = -17.75; education = 5.25; psychology = 17.25. This resulted in statistics of F(1,16) = 37.466; p = 0.00001475 and an effect magnitude of r_effectsize = 0.682.
The manual test gives the same effect size:
lambdas <- rep(c(73, 61, 51, 38), each = 5)
cor(furr_p4$empathy, lambdas)
#> [1] 0.6817294
Let us now run an analysis for within-subjects designs.
Within-Subjects Designs
For within designs the calculations are quite different, but cofad takes
care of the details. We just have to use the within parameters within
and lambda_within instead of the between equivalents. As an example we
use Table 16.5 from Sedlmeier & Renkewitz (2018). Reading ability was
assessed for eight participants under four different conditions. The
hypothesis is that you can read best without music, white noise reduces
your reading ability and music (independently of type) reduces it even
further.
data("sedlmeier_p537")
head(sedlmeier_p537)
#> reading_test participant music
#> 1 27 1 without music
#> 2 25 2 without music
#> 3 30 3 without music
#> 4 29 4 without music
#> 5 30 5 without music
#> 6 33 6 without music
within <- calc_contrast(dv = reading_test, within = music,
lambda_within = c("without music" = 1.25,
"white noise" = 0.25,
"classic" = -0.75,
"jazz" = -0.75),
id = participant, data = sedlmeier_p537)
summary(within)
#> Contrast Analysis Within
#>
#> $Lambdas
#> classic jazz white noise without music
#> -0.75 -0.75 0.25 1.25
#>
#> $tTable
#> mean of L SE df t p(t≥5.269)① 95%CI-lower 95%CI-upper
#> 5.875 1.115 7 5.269 0.000581 3.238 8.512
#>
#> ①The p-value refers to a one-tailed test.
#>
#> $Effects
#>
#> r-contrast 0.687
#> g-contrast 1.863
within
#>
#> We ran a contrast analysis for the following within contrasts: classic = -0.75; jazz = -0.75; white noise = 0.25; without music = 1.25. This resulted in statistics of t(7) = 5.269; p = 0.000581 and an effect magnitude of g_effectsize = 1.863.
You can see that the significance test is just a
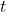 -test and the reported
effect size is referring to a mean comparison
(
-test and the reported
effect size is referring to a mean comparison
(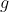 ). (The
). (The
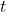 -test is one-tailed,
because contrast analysis has always a specific hypothesis.) When
conducting the analysis by hand, we can see why:
-test is one-tailed,
because contrast analysis has always a specific hypothesis.) When
conducting the analysis by hand, we can see why:
mtr <- matrix(sedlmeier_p537$reading_test, ncol = 4)
lambdas <- c(1.25, 0.25, -0.75, -0.75)
lc1 <- mtr %*% lambdas
t.test(lc1)
#>
#> One Sample t-test
#>
#> data: lc1
#> t = 5.2689, df = 7, p-value = 0.001162
#> alternative hypothesis: true mean is not equal to 0
#> 95 percent confidence interval:
#> 3.238361 8.511639
#> sample estimates:
#> mean of x
#> 5.875
Only the linear combination of the dependent variable and the contrast
weights for each participant is needed. With these values a normal
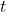 -test against 0 is
conducted. While you can do this manually, using cofad is quicker and it
also gives you more information, such as the different effect sizes.
-test against 0 is
conducted. While you can do this manually, using cofad is quicker and it
also gives you more information, such as the different effect sizes.
Mixed Designs
A mixed design combines between and within factors. In this case cofad
first calculates the linear combination (L-Values) for the within
factor. This new variable serves as the dependent variable for a between
contrast analysis. We will again look at the example presented in
Rosenthal et al. (2000) (see the section graphical user interface). The
cognitive ability of nine children belonging to different age groups
(between) was measured four times (within).
There are two hypotheses:
- cognitive ability linearly increases over time (within)
(
 )
)
- cognitive ability linearly increase over age groups (between)
(
 )
)
Let’s have a look at the data and calculation:
data("rosenthal_tbl53")
head(rosenthal_tbl53)
#> dv between id within
#> 1 3 age8 1 1
#> 2 1 age8 2 1
#> 3 4 age8 3 1
#> 4 4 age10 4 1
#> 5 5 age10 5 1
#> 6 5 age10 6 1
lambda_within <- c("1" = -3, "2" = -1, "3" = 1, "4" = 3)
lambda_between <-c("age8" = -1, "age10" = 0, "age12" = 1)
contr_mx <- calc_contrast(dv = dv,
between = between,
lambda_between = lambda_between,
within = within,
lambda_within = lambda_within,
id = id,
data = rosenthal_tbl53)
contr_mx
#>
#> We ran a contrast analysis for the following between contrasts: age10 = 0; age12 = 1; age8 = -1. This resulted in statistics of F(1,6) = 20.211; p = 0.004123 and an effect magnitude of r_effectsize = 0.871.
The results look like a contrast analysis for between-subject designs.
The summary gives some more details: The effect sizes, within group
means and standard errors of the L-values.
summary(contr_mx)
#> Contrast Analysis Mixed
#>
#> $Lambdas
#> age10 age12 age8
#> 0 1 -1
#>
#> $tTable
#> L df t p(t≥4.496)①
#> 5.333 6 4.496 0.00206
#>
#> ①The p-value refers to a one-tailed test.
#>
#> $FTable
#> SS df MS F p
#> contrast 42.667 1 42.667 20.211 0.00412
#> within 12.667 6 2.111
#> total 56.222 8
#>
#> $Effects
#> effects
#> r_effectsize 0.871
#> r_contrast 0.878
#> r_alerting 0.990
Comparing two hypotheses
With cofad you can also compare two competing hypotheses. As an
example Sedlmeier & Renkewitz (2013) use a fictitious data set on
problem solving skills of boys:
sedlmeier_p525
#> lsg between lambda
#> 1 1 KT -2
#> 2 2 KT -2
#> 3 2 KT -2
#> 4 2 KT -2
#> 5 3 KT -2
#> 6 4 JT 3
#> 7 2 JT 3
#> 8 3 JT 3
#> 9 4 JT 3
#> 10 3 JT 3
#> 11 2 MT -1
#> 12 3 MT -1
#> 13 3 MT -1
#> 14 1 MT -1
#> 15 2 MT -1
Where lsg is the number of solved exercises and the groups are KT = no
training, JT = boys-specific training, MT=girls-specific training. Two
hypotheses are competing:
- -2, 3, -1 (boys benefit from boys-specific training)
- -2, 1, 1 (boys benefit from training, independently of the type of
training)
First, we need to create the difference lambdas:
lambda1 <- c(-2, 3, -1)
lambda2 <- c(-2, 1, 1)
lambda <- lambda_diff(lambda1, lambda2, labels = c("KT", "JT", "MT"))
lambda
#> JT KT MT
#> 0.6816234 0.4883935 -1.1700168
Note that you cannot just subtract the lambdas because their variance
can differ, which has an effect on the test. Instead, you need to
standardize the lambdas first. lambda_diff takes care of this for you.
Now you can run a normal contrast analysis:
ca_competing <- calc_contrast(
dv = lsg,
between = between,
lambda_between = round(lambda, 2),
data = sedlmeier_p525
)
#> lambdas are centered and rounded to 3 digits
summary(ca_competing)
#> Contrast Analysis Between
#>
#> $Lambdas
#> JT KT MT
#> 0.68 0.49 -1.17
#>
#> $tTable
#> L df t p(t≥1.136)①
#> 0.582 12 1.136 0.139
#>
#> ①The p-value refers to a one-tailed test.
#>
#> $FTable
#> SS df MS F p
#> contrast 0.818 1 0.818 1.291 0.278
#> within 7.600 12 0.633
#> total 11.733 14
#>
#> $Effects
#> effects
#> r_effectsize 0.264
#> r_contrast 0.312
#> r_alerting 0.445
ca_competing
#>
#> We ran a contrast analysis for the following between contrasts: JT = 0.68; KT = 0.49; MT = -1.17. This resulted in statistics of F(1,12) = 1.291; p = 0.278 and an effect magnitude of r_effectsize = 0.264.
Here, we rounded the lambdas so that the result is similar to the one in
Sedlmeier & Renkewitz (2013), who found t=1.137 and r_effectsize=0.26.
The effet size is the same. For the t-value, we need to take the root of
the F-value, 1.291, which is 1.136. There is still a slight difference
to the original result of 1.137, which is likely due to rounding errors.
The same also works for within-designs. The reading comprehension data
from above can serve as an example. Reading ability was assessed for
eight participants under four different conditions:
sedlmeier_p537
#> reading_test participant music
#> 1 27 1 without music
#> 2 25 2 without music
#> 3 30 3 without music
#> 4 29 4 without music
#> 5 30 5 without music
#> 6 33 6 without music
#> 7 31 7 without music
#> 8 35 8 without music
#> 9 25 1 white noise
#> 10 26 2 white noise
#> 11 32 3 white noise
#> 12 29 4 white noise
#> 13 28 5 white noise
#> 14 30 6 white noise
#> 15 32 7 white noise
#> 16 34 8 white noise
#> 17 21 1 classic
#> 18 25 2 classic
#> 19 23 3 classic
#> 20 26 4 classic
#> 21 27 5 classic
#> 22 26 6 classic
#> 23 29 7 classic
#> 24 31 8 classic
#> 25 23 1 jazz
#> 26 24 2 jazz
#> 27 24 3 jazz
#> 28 28 4 jazz
#> 29 24 5 jazz
#> 30 26 6 jazz
#> 31 27 7 jazz
#> 32 32 8 jazz
There are two hypotheses:
- 1.25, 0.25, -0.75, -0.75: You can read best without music, white noise
reduces your reading ability and music (independently of type) reduces
it even further.
- 3, -1, -1, -1: Noise of any kind reduces reading ability.
Again, we need to calculate the difference lambdas first:
lambda1 <- c(1.25, 0.25, -0.75, -0.75)
lambda2 <- c(3, -1, -1, -1)
lambda <- lambda_diff(lambda2, lambda1,
labels = c("without music", "white noise", "classic",
"jazz"))
lambda
#> classic jazz white noise without music
#> 0.3271838 0.3271838 -0.8788616 0.2244941
Note that we use lambda2 as the first entry into lambda_diff because
this is how Sedlmeier & Renkewitz (2013) calculated it
(hypothesis2-hypothesis1).
And now the contrast analysis:
contr_wi <- calc_contrast(
dv = reading_test,
within = music,
lambda_within = round(lambda, 2),
id = participant,
data = sedlmeier_p537
)
#> lambdas are centered and rounded to 3 digits
summary(contr_wi)
#> Contrast Analysis Within
#>
#> Your contrast estimate is negative. This means that your data does not reflect the expected direction of your hypothesis specified by the contrast weights (lambdas).
#>
#> $Lambdas
#> classic jazz white noise without music
#> 0.33 0.33 -0.88 0.22
#>
#> $tTable
#> mean of L SE df t p(t≥-3.77)① 95%CI-lower 95%CI-upper
#> -2.2 0.584 7 -3.77 0.997 -3.58 -0.82
#>
#> ①The p-value refers to a one-tailed test.
#>
#> $Effects
#>
#> r-contrast -0.561
#> g-contrast -1.333
contr_wi
#>
#> We ran a contrast analysis for the following within contrasts: classic = 0.33; jazz = 0.33; white noise = -0.88; without music = 0.22. This resulted in statistics of t(7) = -3.77; p = 0.9965 and an effect magnitude of g_effectsize = -1.333. Attention: Contrast fits in the opposite direction!
Sedlmeier & Renkewitz (2013) found a t-value of -3.75 and a g_contrast
of -1.33. Again, there is a slight difference for the t-value when
compared to our calculation, likely due to rounding errors. Further
note, that hypothesis 1 fits better because the statistic and effect are
negative.
Aggregated Data
Sometimes you would like to run a contrast analysis on aggregated data
(e.g. when no raw data is available). If you have the means, standard
deviations and sample sizes for every condition, you can do this with
cofad. For instance, if we take our first example and aggregate it, we
can still run the contrast analysis:
library(dplyr)
furr_agg <- furr_p4 %>%
group_by(major) %>%
summarize(mean = mean(empathy), sd = sd(empathy), n = n())
lambdas = c("psychology" = 1, "education" = -1, "business" = 0, "chemistry" = 0)
calc_contrast_aggregated(mean, sd, n, major, lambdas, furr_agg)
#>
#> We ran a contrast analysis for the following between contrasts: business = 0; chemistry = 0; education = -1; psychology = 1. This resulted in statistics of F(1,16) = 6.154; p = 0.02461 and an effect magnitude of r_effectsize = -0.276. Attention: Contrast fits in the opposite direction!
And the result is indeed the same when compared to the analysis with the
raw data:
ca <- calc_contrast(dv = empathy, between = major,
lambda_between = c("psychology" = 1, "education" = -1,
"business" = 0, "chemistry" = 0),
data = furr_p4)
ca
#>
#> We ran a contrast analysis for the following between contrasts: business = 0; chemistry = 0; education = -1; psychology = 1. This resulted in statistics of F(1,16) = 6.154; p = 0.02461 and an effect magnitude of r_effectsize = -0.276. Attention: Contrast fits in the opposite direction!
Note that this will only work for between-subjects designs.
Testing
The current test coverage for the package stands at 88%. Within cofad,
there exists a Shiny app, subjected to testing through shinytest2.
Regrettably, this testing approach proves less robust, leading to
unpredictable failures specifically on the Windows OS when executed
through GitHub Actions. Consequently, these tests are omitted during
GitHub runs and are exclusively conducted on a local environment.
Issues and Support
If you find any bugs, please use the issue tracker at:
https://github.com/johannes-titz/cofad/issues
If you need answers on how to use the package, drop an e-mail at
johannes at titz.science or johannes.titz at gmail.com
Contributing
Comments and feedback of any kind are very welcome! We will thoroughly
consider every suggestion on how to improve the code, the documentation,
and the presented examples. Even minor things, such as suggestions for
better wording or improving grammar in any part of the package, are more
than welcome.
If you want to make a pull request, please check that you can still
build the package without any errors, warnings, or notes. Overall,
simply stick to the R packages book: https://r-pkgs.org/ and follow
the code style described here: https://style.tidyverse.org/
Acknowledgments
We want to thank Thomas Schäfer and Isabell Winkler for testing cofad
and giving helpful feedback.
References
Furr, R. M. (2004). Interpreting effect sizes in contrast analysis.
*Understanding Statistics*, *3*, 1–25.
Rosenthal, R., & Rosnow, R. L. (1985). *Contrast analysis: Focused
comparisons in the analysis of variance*. Cambridge, England: Cambridge
University Press.
Rosenthal, R., Rosnow, R. L., & Rubin, D. B. (2000). *Contrasts and
Effect Sizes in Behavioral Research: A Correlational Approach*.
Cambridge University Press.
Sedlmeier, P., & Renkewitz, F. (2013). *Forschungsmethoden und Statistik
für Psychologen und Sozialwissenschaftler* (2nd ed.). Hallbergmoos,
Germany: Pearson Studium.
Sedlmeier, P., & Renkewitz, F. (2018). *Forschungsmethoden und Statistik
für Psychologen und Sozialwissenschaftler* (3rd ed.). Hallbergmoos,
Germany: Pearson Studium.
Try the cofad package in your browser
Any scripts or data that you put into this service are public.
cofad documentation built on June 8, 2025, 10:12 a.m.
Cofad User Guide
To cite cofad in publications use:
Titz J. & Burkhardt M. (2021). cofad: An R package and shiny app for contrast analysis. Journal of Open Source Software, 6(67), 3822, https://doi.org/10.21105/joss.03822
A BibTeX entry for LaTeX users is
@article{titz2021,
doi = {10.21105/joss.03822},
url = {https://doi.org/10.21105/joss.03822},
year = {2021},
publisher = {The Open Journal},
volume = {6},
number = {67},
pages = {3822},
author = {Johannes Titz and Markus Burkhardt},
title = {cofad: An R package and shiny app for contrast analysis},
journal = {Journal of Open Source Software} }
Introduction
Cofad is an R package for conducting COntrast analysis in FActorial
Designs like ANOVAs. If contrast analysis was to win a price it would be
the one for the most underestimated, underused statistical technique.
This is unfortunate because in every case a contrast analysis is at
least as good as an ANOVA, but in most cases it is better. Contrast
analysis gets rid off the unspecific omnibus-hypothesis there are
differences somewhere and replaces it with a very specific numerical
hypothesis. Furthermore, contrast analysis focuses on effects instead of
significance. This is expressed doubly: First, there are three different
effect sizes for contrast analysis:
,
and
.
Second, the effect size refers not to the data but to the tested
hypothesis. The larger the effect, the more this speaks for the
hypothesis. One can even compare different hypotheses against each other
(experimentum crucis!) by looking at the effect size for each
hypothesis.
Sounds interesting? Then check out some introductory literature such as Furr (2004), Rosenthal & Rosnow (1985), Rosenthal, Rosnow, & Rubin (2000), or, for the German-speaking audience, Sedlmeier & Renkewitz (2018). Contrast analysis is fairly easy to understand if you know what an ANOVA and a correlation is. In this vignette we assume you are familiar with the basics of contrast analysis and want to apply it to a specific data set. First we show how to install cofad and use the graphical user interface. Then we demonstrate some exemplary analyses for between, within and mixed designs in R.
Installation
Cofad has two components, the plain R package and a shiny-app that offers an intuitive graphical user interface.
If you just want to use the cofad-app, you do not need to install it. Just go to https://cofad.titz.science and use it there. An example data file is loaded when you add /example after the url.
If you prefer the command line interface or want to use the cofad-app locally, install it from CRAN:
install.packages("cofad")
Alternatively, you can also install the development version from github (you need the package devtools for this):
# install.packages("devtools") # uncomment if you do not have devtools installed
devtools::install_github("johannes-titz/cofad")
Now you can load cofad and use it in your R scripts.
You can also run the app:
cofad::run_app()
Using cofad
Before we start: Your data has to be in the long-format (also referred to as narrow or tidy)! If you do not know what this means, please check the short description of the Wikipedia-article: https://en.wikipedia.org/wiki/Wide_and_narrow_data
Graphical-User-Interface
The graphical-user-interface is self-explanatory. Just load your data and drag the variables to the correct position. At the moment you can only read .sav (SPSS) and .csv files.
As an example go to https://cofad.titz.science/example which will load
a data set from Rosenthal et al. (2000) (Table 5.3). The cognitive
ability of nine children belonging to different age groups (between) was
measured four times (within).
There are two hypotheses:
- cognitive ability linearly increases over time (within)
(
)
- cognitive ability linearly increase over age groups (between)
(
)
Now drag the variables to the correct position and set the lambdas accordingly:
 cofad GUI
cofad GUI
The result should look like this:
 cofad GUI
cofad GUI
A mixed design is ideal for testing out the cofad-app. You can now construct a separate within-model by removing the between variable “age”. Then you can construct a separate between-model by removing “time” from within and dragging “age” back into the between panel.
The graphical user interface will suffice for most users, but some will prefer to use the scripting capabilities of R. In the next sections we will look at several script examples for different designs.
Between-Subjects Designs
Let us first load the package:
library(cofad)
Now we need some data and hypotheses. We can simply take the data from Furr (2004), where we have different empathy ratings of students from different majors. This data set is available in the cofad package:
data("furr_p4")
furr_p4
#> empathy major
#> 1 51 psychology
#> 2 56 psychology
#> 3 61 psychology
#> 4 58 psychology
#> 5 54 psychology
#> 6 62 education
#> 7 67 education
#> 8 57 education
#> 9 65 education
#> 10 59 education
#> 11 50 business
#> 12 49 business
#> 13 47 business
#> 14 45 business
#> 15 44 business
#> 16 50 chemistry
#> 17 45 chemistry
#> 18 40 chemistry
#> 19 49 chemistry
#> 20 41 chemistry
Furr states three hypotheses:
- Contrast A: Psychology majors have higher empathy scores than
Education majors
(
).
- Contrast B: Business majors have higher empathy scores than Chemistry
majors
(
).
- Contrast C: On average, Psychology and Education majors have higher
empathy scores than Business and Chemistry majors
(
).
These hypotheses are only mean comparisons, but this is a good way to start. Let’s use cofad to conduct the contrast analysis:
ca <- calc_contrast(dv = empathy, between = major,
lambda_between = c("psychology" = 1, "education" = -1,
"business" = 0, "chemistry" = 0),
data = furr_p4)
ca
#>
#> We ran a contrast analysis for the following between contrasts: business = 0; chemistry = 0; education = -1; psychology = 1. This resulted in statistics of F(1,16) = 6.154; p = 0.02461 and an effect magnitude of r_effectsize = -0.276. Attention: Contrast fits in the opposite direction!
The print method shows some basic information that can be directly used in a publication. With the summary method some more details are shown:
summary(ca)
#> Contrast Analysis Between
#>
#> Your contrast estimate is negative. This means that your data does not reflect the expected direction of your hypothesis specified by the contrast weights (lambdas).
#>
#> $Lambdas
#> business chemistry education psychology
#> 0 0 -1 1
#>
#> $tTable
#> L df t p(t≥-2.481)①
#> -6 16 -2.481 0.988
#>
#> ①The p-value refers to a one-tailed test.
#>
#> $FTable
#> SS df MS F p
#> contrast 90 1 90.000 6.154 0.0246
#> within 234 16 14.625
#> total 1179 19
#>
#> $Effects
#> effects
#> r_effectsize -0.276
#> r_contrast -0.527
#> r_alerting -0.309
From this table,
is probably the most useful statistic. It is just the correlation
between the lambdas and the dependent variable, which can also be
calculated by hand:
lambdas <- rep(c(1, -1, 0, 0), each = 5)
cor(furr_p4$empathy, lambdas)
#> [1] -0.2762895
As you can see, the effect is negative and cofad also warns the user
that the contrast fits in the opposite direction. This is a big failure
for the hypothesis and indicates substantial problems in theorizing.
The other two hypotheses can be tested accordingly:
ca <- calc_contrast(dv = empathy, between = major,
lambda_between = c("psychology" = 0, "education" = 0,
"business" = 1, "chemistry" = -1),
data = furr_p4)
ca
#>
#> We ran a contrast analysis for the following between contrasts: business = 1; chemistry = -1; education = 0; psychology = 0. This resulted in statistics of F(1,16) = 0.684; p = 0.4205 and an effect magnitude of r_effectsize = 0.092.
ca <- calc_contrast(dv = empathy, between = major,
lambda_between = c("psychology" = 1, "education" = 1,
"business" = -1, "chemistry" = -1),
data = furr_p4)
ca
#>
#> We ran a contrast analysis for the following between contrasts: business = -1; chemistry = -1; education = 1; psychology = 1. This resulted in statistics of F(1,16) = 57.778; p = 0.00000107 and an effect magnitude of r_effectsize = 0.847.
When you compare the numbers to the ones presented in Furr (2004), you will find the same result, except that Furr (2004) uses t-values and the p-values are halved. This is because in contrast analysis you can always test one-sided. The assumption is that your lambdas covariate positively with the mean values, not that they either covariate positively or negatively. Thus, you can always halve the p-value from the F-Test.
Now, imagine we have a more fun hypothesis and not just mean differences. From an elaborate theory we could derive that the means should be 73, 61, 51 and 38. We can test this with cofad directly because cofad will center the lambdas (the mean of the lambdas has to be 0):
ca <- calc_contrast(dv = empathy, between = major,
lambda_between = c("psychology" = 73, "education" = 61,
"business" = 51, "chemistry" = 38),
data = furr_p4)
#> lambdas are centered and rounded to 3 digits
ca
#>
#> We ran a contrast analysis for the following between contrasts: business = -4.75; chemistry = -17.75; education = 5.25; psychology = 17.25. This resulted in statistics of F(1,16) = 37.466; p = 0.00001475 and an effect magnitude of r_effectsize = 0.682.
The manual test gives the same effect size:
lambdas <- rep(c(73, 61, 51, 38), each = 5)
cor(furr_p4$empathy, lambdas)
#> [1] 0.6817294
Let us now run an analysis for within-subjects designs.
Within-Subjects Designs
For within designs the calculations are quite different, but cofad takes care of the details. We just have to use the within parameters within and lambda_within instead of the between equivalents. As an example we use Table 16.5 from Sedlmeier & Renkewitz (2018). Reading ability was assessed for eight participants under four different conditions. The hypothesis is that you can read best without music, white noise reduces your reading ability and music (independently of type) reduces it even further.
data("sedlmeier_p537")
head(sedlmeier_p537)
#> reading_test participant music
#> 1 27 1 without music
#> 2 25 2 without music
#> 3 30 3 without music
#> 4 29 4 without music
#> 5 30 5 without music
#> 6 33 6 without music
within <- calc_contrast(dv = reading_test, within = music,
lambda_within = c("without music" = 1.25,
"white noise" = 0.25,
"classic" = -0.75,
"jazz" = -0.75),
id = participant, data = sedlmeier_p537)
summary(within)
#> Contrast Analysis Within
#>
#> $Lambdas
#> classic jazz white noise without music
#> -0.75 -0.75 0.25 1.25
#>
#> $tTable
#> mean of L SE df t p(t≥5.269)① 95%CI-lower 95%CI-upper
#> 5.875 1.115 7 5.269 0.000581 3.238 8.512
#>
#> ①The p-value refers to a one-tailed test.
#>
#> $Effects
#>
#> r-contrast 0.687
#> g-contrast 1.863
within
#>
#> We ran a contrast analysis for the following within contrasts: classic = -0.75; jazz = -0.75; white noise = 0.25; without music = 1.25. This resulted in statistics of t(7) = 5.269; p = 0.000581 and an effect magnitude of g_effectsize = 1.863.
You can see that the significance test is just a
-test and the reported
effect size is referring to a mean comparison
(
). (The
-test is one-tailed,
because contrast analysis has always a specific hypothesis.) When
conducting the analysis by hand, we can see why:
mtr <- matrix(sedlmeier_p537$reading_test, ncol = 4)
lambdas <- c(1.25, 0.25, -0.75, -0.75)
lc1 <- mtr %*% lambdas
t.test(lc1)
#>
#> One Sample t-test
#>
#> data: lc1
#> t = 5.2689, df = 7, p-value = 0.001162
#> alternative hypothesis: true mean is not equal to 0
#> 95 percent confidence interval:
#> 3.238361 8.511639
#> sample estimates:
#> mean of x
#> 5.875
Only the linear combination of the dependent variable and the contrast
weights for each participant is needed. With these values a normal
-test against 0 is
conducted. While you can do this manually, using cofad is quicker and it
also gives you more information, such as the different effect sizes.
Mixed Designs
A mixed design combines between and within factors. In this case cofad first calculates the linear combination (L-Values) for the within factor. This new variable serves as the dependent variable for a between contrast analysis. We will again look at the example presented in Rosenthal et al. (2000) (see the section graphical user interface). The cognitive ability of nine children belonging to different age groups (between) was measured four times (within).
There are two hypotheses:
- cognitive ability linearly increases over time (within)
(
)
- cognitive ability linearly increase over age groups (between)
(
)
Let’s have a look at the data and calculation:
data("rosenthal_tbl53")
head(rosenthal_tbl53)
#> dv between id within
#> 1 3 age8 1 1
#> 2 1 age8 2 1
#> 3 4 age8 3 1
#> 4 4 age10 4 1
#> 5 5 age10 5 1
#> 6 5 age10 6 1
lambda_within <- c("1" = -3, "2" = -1, "3" = 1, "4" = 3)
lambda_between <-c("age8" = -1, "age10" = 0, "age12" = 1)
contr_mx <- calc_contrast(dv = dv,
between = between,
lambda_between = lambda_between,
within = within,
lambda_within = lambda_within,
id = id,
data = rosenthal_tbl53)
contr_mx
#>
#> We ran a contrast analysis for the following between contrasts: age10 = 0; age12 = 1; age8 = -1. This resulted in statistics of F(1,6) = 20.211; p = 0.004123 and an effect magnitude of r_effectsize = 0.871.
The results look like a contrast analysis for between-subject designs. The summary gives some more details: The effect sizes, within group means and standard errors of the L-values.
summary(contr_mx)
#> Contrast Analysis Mixed
#>
#> $Lambdas
#> age10 age12 age8
#> 0 1 -1
#>
#> $tTable
#> L df t p(t≥4.496)①
#> 5.333 6 4.496 0.00206
#>
#> ①The p-value refers to a one-tailed test.
#>
#> $FTable
#> SS df MS F p
#> contrast 42.667 1 42.667 20.211 0.00412
#> within 12.667 6 2.111
#> total 56.222 8
#>
#> $Effects
#> effects
#> r_effectsize 0.871
#> r_contrast 0.878
#> r_alerting 0.990
Comparing two hypotheses
With cofad you can also compare two competing hypotheses. As an
example Sedlmeier & Renkewitz (2013) use a fictitious data set on
problem solving skills of boys:
sedlmeier_p525
#> lsg between lambda
#> 1 1 KT -2
#> 2 2 KT -2
#> 3 2 KT -2
#> 4 2 KT -2
#> 5 3 KT -2
#> 6 4 JT 3
#> 7 2 JT 3
#> 8 3 JT 3
#> 9 4 JT 3
#> 10 3 JT 3
#> 11 2 MT -1
#> 12 3 MT -1
#> 13 3 MT -1
#> 14 1 MT -1
#> 15 2 MT -1
Where lsg is the number of solved exercises and the groups are KT = no training, JT = boys-specific training, MT=girls-specific training. Two hypotheses are competing:
- -2, 3, -1 (boys benefit from boys-specific training)
- -2, 1, 1 (boys benefit from training, independently of the type of training)
First, we need to create the difference lambdas:
lambda1 <- c(-2, 3, -1)
lambda2 <- c(-2, 1, 1)
lambda <- lambda_diff(lambda1, lambda2, labels = c("KT", "JT", "MT"))
lambda
#> JT KT MT
#> 0.6816234 0.4883935 -1.1700168
Note that you cannot just subtract the lambdas because their variance
can differ, which has an effect on the test. Instead, you need to
standardize the lambdas first. lambda_diff takes care of this for you.
Now you can run a normal contrast analysis:
ca_competing <- calc_contrast(
dv = lsg,
between = between,
lambda_between = round(lambda, 2),
data = sedlmeier_p525
)
#> lambdas are centered and rounded to 3 digits
summary(ca_competing)
#> Contrast Analysis Between
#>
#> $Lambdas
#> JT KT MT
#> 0.68 0.49 -1.17
#>
#> $tTable
#> L df t p(t≥1.136)①
#> 0.582 12 1.136 0.139
#>
#> ①The p-value refers to a one-tailed test.
#>
#> $FTable
#> SS df MS F p
#> contrast 0.818 1 0.818 1.291 0.278
#> within 7.600 12 0.633
#> total 11.733 14
#>
#> $Effects
#> effects
#> r_effectsize 0.264
#> r_contrast 0.312
#> r_alerting 0.445
ca_competing
#>
#> We ran a contrast analysis for the following between contrasts: JT = 0.68; KT = 0.49; MT = -1.17. This resulted in statistics of F(1,12) = 1.291; p = 0.278 and an effect magnitude of r_effectsize = 0.264.
Here, we rounded the lambdas so that the result is similar to the one in Sedlmeier & Renkewitz (2013), who found t=1.137 and r_effectsize=0.26. The effet size is the same. For the t-value, we need to take the root of the F-value, 1.291, which is 1.136. There is still a slight difference to the original result of 1.137, which is likely due to rounding errors.
The same also works for within-designs. The reading comprehension data from above can serve as an example. Reading ability was assessed for eight participants under four different conditions:
sedlmeier_p537
#> reading_test participant music
#> 1 27 1 without music
#> 2 25 2 without music
#> 3 30 3 without music
#> 4 29 4 without music
#> 5 30 5 without music
#> 6 33 6 without music
#> 7 31 7 without music
#> 8 35 8 without music
#> 9 25 1 white noise
#> 10 26 2 white noise
#> 11 32 3 white noise
#> 12 29 4 white noise
#> 13 28 5 white noise
#> 14 30 6 white noise
#> 15 32 7 white noise
#> 16 34 8 white noise
#> 17 21 1 classic
#> 18 25 2 classic
#> 19 23 3 classic
#> 20 26 4 classic
#> 21 27 5 classic
#> 22 26 6 classic
#> 23 29 7 classic
#> 24 31 8 classic
#> 25 23 1 jazz
#> 26 24 2 jazz
#> 27 24 3 jazz
#> 28 28 4 jazz
#> 29 24 5 jazz
#> 30 26 6 jazz
#> 31 27 7 jazz
#> 32 32 8 jazz
There are two hypotheses:
- 1.25, 0.25, -0.75, -0.75: You can read best without music, white noise reduces your reading ability and music (independently of type) reduces it even further.
- 3, -1, -1, -1: Noise of any kind reduces reading ability.
Again, we need to calculate the difference lambdas first:
lambda1 <- c(1.25, 0.25, -0.75, -0.75)
lambda2 <- c(3, -1, -1, -1)
lambda <- lambda_diff(lambda2, lambda1,
labels = c("without music", "white noise", "classic",
"jazz"))
lambda
#> classic jazz white noise without music
#> 0.3271838 0.3271838 -0.8788616 0.2244941
Note that we use lambda2 as the first entry into lambda_diff because
this is how Sedlmeier & Renkewitz (2013) calculated it
(hypothesis2-hypothesis1).
And now the contrast analysis:
contr_wi <- calc_contrast(
dv = reading_test,
within = music,
lambda_within = round(lambda, 2),
id = participant,
data = sedlmeier_p537
)
#> lambdas are centered and rounded to 3 digits
summary(contr_wi)
#> Contrast Analysis Within
#>
#> Your contrast estimate is negative. This means that your data does not reflect the expected direction of your hypothesis specified by the contrast weights (lambdas).
#>
#> $Lambdas
#> classic jazz white noise without music
#> 0.33 0.33 -0.88 0.22
#>
#> $tTable
#> mean of L SE df t p(t≥-3.77)① 95%CI-lower 95%CI-upper
#> -2.2 0.584 7 -3.77 0.997 -3.58 -0.82
#>
#> ①The p-value refers to a one-tailed test.
#>
#> $Effects
#>
#> r-contrast -0.561
#> g-contrast -1.333
contr_wi
#>
#> We ran a contrast analysis for the following within contrasts: classic = 0.33; jazz = 0.33; white noise = -0.88; without music = 0.22. This resulted in statistics of t(7) = -3.77; p = 0.9965 and an effect magnitude of g_effectsize = -1.333. Attention: Contrast fits in the opposite direction!
Sedlmeier & Renkewitz (2013) found a t-value of -3.75 and a g_contrast of -1.33. Again, there is a slight difference for the t-value when compared to our calculation, likely due to rounding errors. Further note, that hypothesis 1 fits better because the statistic and effect are negative.
Aggregated Data
Sometimes you would like to run a contrast analysis on aggregated data (e.g. when no raw data is available). If you have the means, standard deviations and sample sizes for every condition, you can do this with cofad. For instance, if we take our first example and aggregate it, we can still run the contrast analysis:
library(dplyr)
furr_agg <- furr_p4 %>%
group_by(major) %>%
summarize(mean = mean(empathy), sd = sd(empathy), n = n())
lambdas = c("psychology" = 1, "education" = -1, "business" = 0, "chemistry" = 0)
calc_contrast_aggregated(mean, sd, n, major, lambdas, furr_agg)
#>
#> We ran a contrast analysis for the following between contrasts: business = 0; chemistry = 0; education = -1; psychology = 1. This resulted in statistics of F(1,16) = 6.154; p = 0.02461 and an effect magnitude of r_effectsize = -0.276. Attention: Contrast fits in the opposite direction!
And the result is indeed the same when compared to the analysis with the raw data:
ca <- calc_contrast(dv = empathy, between = major,
lambda_between = c("psychology" = 1, "education" = -1,
"business" = 0, "chemistry" = 0),
data = furr_p4)
ca
#>
#> We ran a contrast analysis for the following between contrasts: business = 0; chemistry = 0; education = -1; psychology = 1. This resulted in statistics of F(1,16) = 6.154; p = 0.02461 and an effect magnitude of r_effectsize = -0.276. Attention: Contrast fits in the opposite direction!
Note that this will only work for between-subjects designs.
Testing
The current test coverage for the package stands at 88%. Within cofad,
there exists a Shiny app, subjected to testing through shinytest2.
Regrettably, this testing approach proves less robust, leading to
unpredictable failures specifically on the Windows OS when executed
through GitHub Actions. Consequently, these tests are omitted during
GitHub runs and are exclusively conducted on a local environment.
Issues and Support
If you find any bugs, please use the issue tracker at:
https://github.com/johannes-titz/cofad/issues
If you need answers on how to use the package, drop an e-mail at johannes at titz.science or johannes.titz at gmail.com
Contributing
Comments and feedback of any kind are very welcome! We will thoroughly consider every suggestion on how to improve the code, the documentation, and the presented examples. Even minor things, such as suggestions for better wording or improving grammar in any part of the package, are more than welcome.
If you want to make a pull request, please check that you can still build the package without any errors, warnings, or notes. Overall, simply stick to the R packages book: https://r-pkgs.org/ and follow the code style described here: https://style.tidyverse.org/
Acknowledgments
We want to thank Thomas Schäfer and Isabell Winkler for testing cofad and giving helpful feedback.
References
Try the cofad package in your browser
Any scripts or data that you put into this service are public.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
