collapse and sf
In collapse: Advanced and Fast Data Transformation
This short vignette focuses on using collapse with the popular sf package by Edzer Pebesma. It shows that collapse supports easy manipulation of sf data frames, at computation speeds far above dplyr.
collapse v1.6.0 added internal support for sf data frames by having most essential functions (e.g., fselect/gv, fsubset/ss, fgroup_by, findex_by, qsu, descr, varying, funique, roworder, rsplit, fcompute, ...) internally handle the geometry column.
To demonstrate this, we can load a test dataset provided by sf:
library(collapse)
library(sf)
nc <- st_read(system.file("shape/nc.shp", package = "sf"), quiet = TRUE)
options(sf_max_print = 3)
nc
# Simple feature collection with 100 features and 14 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79 SID79
# 1 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 1 10 1364 0
# 2 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 0 10 542 3
# 3 0.143 1.630 1828 1828 Surry 37171 37171 86 3188 5 208 3616 6
# NWBIR79 geometry
# 1 19 MULTIPOLYGON (((-81.47276 3...
# 2 12 MULTIPOLYGON (((-81.23989 3...
# 3 260 MULTIPOLYGON (((-80.45634 3...
Summarising sf Data Frames
Computing summary statistics on sf data frames automatically excludes the 'geometry' column:
# Which columns have at least 2 non-missing distinct values
varying(nc)
# AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74
# TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
# NWBIR74 BIR79 SID79 NWBIR79
# TRUE TRUE TRUE TRUE
# Quick summary stats
qsu(nc)
# N Mean SD Min Max
# AREA 100 0.1263 0.0492 0.042 0.241
# PERIMETER 100 1.673 0.4823 0.999 3.64
# CNTY_ 100 1985.96 106.5166 1825 2241
# CNTY_ID 100 1985.96 106.5166 1825 2241
# NAME 100 - - - -
# FIPS 100 - - - -
# FIPSNO 100 37100 58.023 37001 37199
# CRESS_ID 100 50.5 29.0115 1 100
# BIR74 100 3299.62 3848.1651 248 21588
# SID74 100 6.67 7.7812 0 44
# NWBIR74 100 1050.81 1432.9117 1 8027
# BIR79 100 4223.92 5179.4582 319 30757
# SID79 100 8.36 9.4319 0 57
# NWBIR79 100 1352.81 1975.9988 3 11631
# Detailed statistics description of each column
descr(nc)
# Dataset: nc, 14 Variables, N = 100
# ----------------------------------------------------------------------------------------------------
# AREA (numeric):
# Statistics
# N Ndist Mean SD Min Max Skew Kurt
# 100 77 0.13 0.05 0.04 0.24 0.48 2.5
# Quantiles
# 1% 5% 10% 25% 50% 75% 90% 95% 99%
# 0.04 0.06 0.06 0.09 0.12 0.15 0.2 0.21 0.24
# ----------------------------------------------------------------------------------------------------
# PERIMETER (numeric):
# Statistics
# N Ndist Mean SD Min Max Skew Kurt
# 100 96 1.67 0.48 1 3.64 1.48 5.95
# Quantiles
# 1% 5% 10% 25% 50% 75% 90% 95% 99%
# 1 1.09 1.19 1.32 1.61 1.86 2.2 2.72 3.2
# ----------------------------------------------------------------------------------------------------
# CNTY_ (numeric):
# Statistics
# N Ndist Mean SD Min Max Skew Kurt
# 100 100 1985.96 106.52 1825 2241 0.26 2.32
# Quantiles
# 1% 5% 10% 25% 50% 75% 90% 95% 99%
# 1826.98 1832.95 1837.9 1902.25 1982 2067.25 2110 2156.3 2238.03
# ----------------------------------------------------------------------------------------------------
# CNTY_ID (numeric):
# Statistics
# N Ndist Mean SD Min Max Skew Kurt
# 100 100 1985.96 106.52 1825 2241 0.26 2.32
# Quantiles
# 1% 5% 10% 25% 50% 75% 90% 95% 99%
# 1826.98 1832.95 1837.9 1902.25 1982 2067.25 2110 2156.3 2238.03
# ----------------------------------------------------------------------------------------------------
# NAME (character):
# Statistics
# N Ndist
# 100 100
# Table
# Freq Perc
# Ashe 1 1
# Alleghany 1 1
# Surry 1 1
# Currituck 1 1
# Northampton 1 1
# Hertford 1 1
# Camden 1 1
# Gates 1 1
# Warren 1 1
# Stokes 1 1
# Caswell 1 1
# Rockingham 1 1
# Granville 1 1
# Person 1 1
# ... 86 Others 86 86
#
# Summary of Table Frequencies
# Min. 1st Qu. Median Mean 3rd Qu. Max.
# 1 1 1 1 1 1
# ----------------------------------------------------------------------------------------------------
# FIPS (character):
# Statistics
# N Ndist
# 100 100
# Table
# Freq Perc
# 37009 1 1
# 37005 1 1
# 37171 1 1
# 37053 1 1
# 37131 1 1
# 37091 1 1
# 37029 1 1
# 37073 1 1
# 37185 1 1
# 37169 1 1
# 37033 1 1
# 37157 1 1
# 37077 1 1
# 37145 1 1
# ... 86 Others 86 86
#
# Summary of Table Frequencies
# Min. 1st Qu. Median Mean 3rd Qu. Max.
# 1 1 1 1 1 1
# ----------------------------------------------------------------------------------------------------
# FIPSNO (numeric):
# Statistics
# N Ndist Mean SD Min Max Skew Kurt
# 100 100 37100 58.02 37001 37199 -0 1.8
# Quantiles
# 1% 5% 10% 25% 50% 75% 90% 95% 99%
# 37002.98 37010.9 37020.8 37050.5 37100 37149.5 37179.2 37189.1 37197.02
# ----------------------------------------------------------------------------------------------------
# CRESS_ID (integer):
# Statistics
# N Ndist Mean SD Min Max Skew Kurt
# 100 100 50.5 29.01 1 100 0 1.8
# Quantiles
# 1% 5% 10% 25% 50% 75% 90% 95% 99%
# 1.99 5.95 10.9 25.75 50.5 75.25 90.1 95.05 99.01
# ----------------------------------------------------------------------------------------------------
# BIR74 (numeric):
# Statistics
# N Ndist Mean SD Min Max Skew Kurt
# 100 100 3299.62 3848.17 248 21588 2.79 11.79
# Quantiles
# 1% 5% 10% 25% 50% 75% 90% 95% 99%
# 283.64 419.75 531.8 1077 2180.5 3936 6725.7 11193 20378.22
# ----------------------------------------------------------------------------------------------------
# SID74 (numeric):
# Statistics
# N Ndist Mean SD Min Max Skew Kurt
# 100 23 6.67 7.78 0 44 2.44 10.28
# Quantiles
# 1% 5% 10% 25% 50% 75% 90% 95% 99%
# 0 0 0 2 4 8.25 15.1 18.25 38.06
# ----------------------------------------------------------------------------------------------------
# NWBIR74 (numeric):
# Statistics
# N Ndist Mean SD Min Max Skew Kurt
# 100 93 1050.81 1432.91 1 8027 2.83 11.84
# Quantiles
# 1% 5% 10% 25% 50% 75% 90% 95% 99%
# 1 9.95 39.2 190 697.5 1168.5 2231.8 3942.9 7052.84
# ----------------------------------------------------------------------------------------------------
# BIR79 (numeric):
# Statistics
# N Ndist Mean SD Min Max Skew Kurt
# 100 100 4223.92 5179.46 319 30757 2.99 13.1
# Quantiles
# 1% 5% 10% 25% 50% 75% 90% 95% 99%
# 349.69 539.3 675.7 1336.25 2636 4889 8313 14707.45 26413.87
# ----------------------------------------------------------------------------------------------------
# SID79 (numeric):
# Statistics
# N Ndist Mean SD Min Max Skew Kurt
# 100 28 8.36 9.43 0 57 2.28 9.88
# Quantiles
# 1% 5% 10% 25% 50% 75% 90% 95% 99%
# 0 0 1 2 5 10.25 21 26 38.19
# ----------------------------------------------------------------------------------------------------
# NWBIR79 (numeric):
# Statistics
# N Ndist Mean SD Min Max Skew Kurt
# 100 98 1352.81 1976 3 11631 3.18 14.45
# Quantiles
# 1% 5% 10% 25% 50% 75% 90% 95% 99%
# 3.99 11.9 44.7 250.5 874.5 1406.75 2987.9 5090.5 10624.17
# ----------------------------------------------------------------------------------------------------
Selecting Columns and Subsetting
We can select columns from the sf data frame without having to worry about taking along 'geometry':
# Selecting a sequence of columns
fselect(nc, AREA, NAME:FIPSNO)
# Simple feature collection with 100 features and 4 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# AREA NAME FIPS FIPSNO geometry
# 1 0.114 Ashe 37009 37009 MULTIPOLYGON (((-81.47276 3...
# 2 0.061 Alleghany 37005 37005 MULTIPOLYGON (((-81.23989 3...
# 3 0.143 Surry 37171 37171 MULTIPOLYGON (((-80.45634 3...
# Same using standard evaluation (gv is a shorthand for get_vars())
gv(nc, c("AREA", "NAME", "FIPS", "FIPSNO"))
# Simple feature collection with 100 features and 4 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# AREA NAME FIPS FIPSNO geometry
# 1 0.114 Ashe 37009 37009 MULTIPOLYGON (((-81.47276 3...
# 2 0.061 Alleghany 37005 37005 MULTIPOLYGON (((-81.23989 3...
# 3 0.143 Surry 37171 37171 MULTIPOLYGON (((-80.45634 3...
The same applies to subsetting rows (and columns):
# A fast and enhanced version of base::subset
fsubset(nc, AREA > fmean(AREA), AREA, NAME:FIPSNO)
# Simple feature collection with 44 features and 4 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# AREA NAME FIPS FIPSNO geometry
# 1 0.143 Surry 37171 37171 MULTIPOLYGON (((-80.45634 3...
# 2 0.153 Northampton 37131 37131 MULTIPOLYGON (((-77.21767 3...
# 3 0.153 Rockingham 37157 37157 MULTIPOLYGON (((-79.53051 3...
# A fast version of `[` (where i is used and optionally j)
ss(nc, 1:10, c("AREA", "NAME", "FIPS", "FIPSNO"))
# Simple feature collection with 10 features and 4 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# AREA NAME FIPS FIPSNO geometry
# 1 0.114 Ashe 37009 37009 MULTIPOLYGON (((-81.47276 3...
# 2 0.061 Alleghany 37005 37005 MULTIPOLYGON (((-81.23989 3...
# 3 0.143 Surry 37171 37171 MULTIPOLYGON (((-80.45634 3...
This is significantly faster than using [, base::subset(), dplyr::select() or dplyr::filter():
library(microbenchmark)
library(dplyr)
# Selecting columns
microbenchmark(collapse = fselect(nc, AREA, NAME:FIPSNO),
dplyr = select(nc, AREA, NAME:FIPSNO),
collapse2 = gv(nc, c("AREA", "NAME", "FIPS", "FIPSNO")),
sf = nc[c("AREA", "NAME", "FIPS", "FIPSNO")])
# Unit: microseconds
# expr min lq mean median uq max neval
# collapse 3.034 3.9565 5.19429 5.1865 5.6990 22.878 100
# dplyr 431.279 452.2915 505.29015 466.3750 493.8450 3356.342 100
# collapse2 2.665 3.4850 4.59610 4.4075 5.0635 14.391 100
# sf 105.165 114.1235 120.39732 118.0390 124.9270 156.497 100
# Subsetting
microbenchmark(collapse = fsubset(nc, AREA > fmean(AREA), AREA, NAME:FIPSNO),
dplyr = select(nc, AREA, NAME:FIPSNO) |> filter(AREA > fmean(AREA)),
collapse2 = ss(nc, 1:10, c("AREA", "NAME", "FIPS", "FIPSNO")),
sf = nc[1:10, c("AREA", "NAME", "FIPS", "FIPSNO")])
# Unit: microseconds
# expr min lq mean median uq max neval
# collapse 9.676 11.5825 15.01707 14.4730 16.8920 30.463 100
# dplyr 890.643 917.6415 1055.40970 941.7085 1009.7890 5546.685 100
# collapse2 2.829 3.5465 5.40585 4.8995 6.4165 20.541 100
# sf 176.997 187.6160 202.72286 200.7565 210.8220 340.464 100
However, collapse functions don't subset the 'agr' attribute on selecting columns, which (if specified) relates columns (attributes) to the geometry, and also don't modify the 'bbox' attribute giving the overall boundaries of a set of geometries when subsetting the sf data frame. Keeping the full 'agr' attribute is not problematic for all practical purposes, but not changing 'bbox' upon subsetting may lead to too large margins when plotting the geometries of a subset sf data frame.
One way to to change this is calling st_make_valid() on the subset frame; but st_make_valid() is very expensive, thus unless the subset frame is very small, it is better to use [, base::subset() or dplyr::filter() in cases where the bounding box size matters.
Aggregation and Grouping
The flexibility and speed of collap() for aggregation can be used on sf data frames. A separate method for sf objects was not considered necessary as one can simply aggregate the geometry column using st_union():
# Aggregating by variable SID74 using the median for numeric and the mode for categorical columns
collap(nc, ~ SID74, custom = list(fmedian = is.numeric,
fmode = is.character,
st_union = "geometry")) # or use is.list to fetch the geometry
# Simple feature collection with 23 features and 15 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 SID74 NWBIR74 BIR79
# 1 0.0780 1.3070 1950.0 1950.0 Alleghany 37005 37073 37.0 487 0 0 40.0 594.0
# 2 0.0810 1.2880 1887.0 1887.0 Ashe 37009 37137 69.0 751 1 1 148.0 899.0
# 3 0.1225 1.6435 1959.5 1959.5 Caswell 37033 37078 39.5 1271 2 2 382.5 1676.5
# SID79 NWBIR79 geometry
# 1 1 45 MULTIPOLYGON (((-83.69563 3...
# 2 1 176 MULTIPOLYGON (((-80.02406 3...
# 3 2 452 MULTIPOLYGON (((-77.16129 3...
sf data frames can also be grouped and then aggregated using fsummarise():
nc |> fgroup_by(SID74)
# Simple feature collection with 100 features and 14 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79 SID79
# 1 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 1 10 1364 0
# 2 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 0 10 542 3
# 3 0.143 1.630 1828 1828 Surry 37171 37171 86 3188 5 208 3616 6
# NWBIR79 geometry
# 1 19 MULTIPOLYGON (((-81.47276 3...
# 2 12 MULTIPOLYGON (((-81.23989 3...
# 3 260 MULTIPOLYGON (((-80.45634 3...
#
# Grouped by: SID74 [23 | 4 (4) 1-13]
nc |>
fgroup_by(SID74) |>
fsummarise(AREA_Ag = fsum(AREA),
Perimeter_Ag = fmedian(PERIMETER),
geometry = st_union(geometry))
# Simple feature collection with 23 features and 3 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# SID74 AREA_Ag Perimeter_Ag geometry
# 1 0 1.103 1.3070 MULTIPOLYGON (((-83.69563 3...
# 2 1 0.914 1.2880 MULTIPOLYGON (((-80.02406 3...
# 3 2 1.047 1.6435 MULTIPOLYGON (((-77.16129 3...
Typically most of the time in aggregation is consumed by st_union() so that the speed of collapse does not really become visible on most datasets. A faster alternative is to use geos (sf backend for planar geometries) or s2 (sf backend for spherical geometries) directly:
# Using s2 backend: sensible for larger tasks
nc |>
fmutate(geometry = s2::as_s2_geography(geometry)) |>
fgroup_by(SID74) |>
fsummarise(AREA_Ag = fsum(AREA),
Perimeter_Ag = fmedian(PERIMETER),
geometry = s2::s2_union_agg(geometry)) |>
fmutate(geometry = st_as_sfc(geometry))
# Simple feature collection with 23 features and 3 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: WGS 84
# First 3 features:
# SID74 AREA_Ag Perimeter_Ag geometry
# 1 0 1.103 1.3070 MULTIPOLYGON (((-83.69563 3...
# 2 1 0.914 1.2880 MULTIPOLYGON (((-80.02406 3...
# 3 2 1.047 1.6435 MULTIPOLYGON (((-77.16129 3...
In general, also upon aggregation with collapse, functions st_as_sfc(), st_as_sf(), or, in the worst case, st_make_valid(), may need to be invoked to ensure valid sf object output. Functions collap() and fsummarise() are attribute preserving but do not give special regard to geometry columns.
One exception that both avoids the high cost of spatial functions in aggregation and any need for ex-post conversion/validation is aggregating spatial panel data over the time-dimension. Such panels can quickly be aggregated using ffirst() or flast() to aggregate the geometry:
# Creating a panel-dataset by simply duplicating nc for 2 different years
pnc <- rowbind(`2000` = nc, `2001` = nc, idcol = "Year") |> as_integer_factor()
pnc
# Simple feature collection with 200 features and 15 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# Year AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79
# 1 2000 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 1 10 1364
# 2 2000 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 0 10 542
# 3 2000 0.143 1.630 1828 1828 Surry 37171 37171 86 3188 5 208 3616
# SID79 NWBIR79 geometry
# 1 0 19 MULTIPOLYGON (((-81.47276 3...
# 2 3 12 MULTIPOLYGON (((-81.23989 3...
# 3 6 260 MULTIPOLYGON (((-80.45634 3...
# Aggregating by NAME, using the last value for all categorical data
collap(pnc, ~ NAME, fmedian, catFUN = flast, cols = -1L)
# Simple feature collection with 100 features and 15 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# AREA PERIMETER CNTY_ CNTY_ID NAME NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79
# 1 0.111 1.392 1904 1904 Alamance Alamance 37001 37001 1 4672 13 1243 5767
# 2 0.066 1.070 1950 1950 Alexander Alexander 37003 37003 2 1333 0 128 1683
# 3 0.061 1.231 1827 1827 Alleghany Alleghany 37005 37005 3 487 0 10 542
# SID79 NWBIR79 geometry
# 1 11 1397 MULTIPOLYGON (((-79.24619 3...
# 2 2 150 MULTIPOLYGON (((-81.10889 3...
# 3 3 12 MULTIPOLYGON (((-81.23989 3...
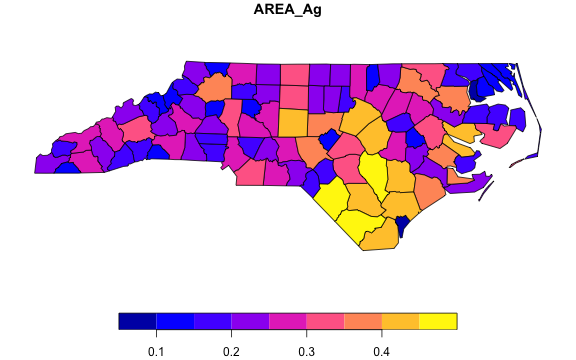
# Using fsummarise to aggregate just two variables and the geometry
pnc_ag <- pnc |>
fgroup_by(NAME) |>
fsummarise(AREA_Ag = fsum(AREA),
Perimeter_Ag = fmedian(PERIMETER),
geometry = flast(geometry))
# The geometry is still valid... (slt = shorthand for fselect)
plot(slt(pnc_ag, AREA_Ag))
Indexing
sf data frames can also become indexed frames (spatio-temporal panels):
pnc <- pnc |> findex_by(CNTY_ID, Year)
pnc
# Simple feature collection with 200 features and 15 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# Year AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79
# 1 2000 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 1 10 1364
# 2 2000 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 0 10 542
# 3 2000 0.143 1.630 1828 1828 Surry 37171 37171 86 3188 5 208 3616
# SID79 NWBIR79 geometry
# 1 0 19 MULTIPOLYGON (((-81.47276 3...
# 2 3 12 MULTIPOLYGON (((-81.23989 3...
# 3 6 260 MULTIPOLYGON (((-80.45634 3...
#
# Indexed by: CNTY_ID [100] | Year [2]
qsu(pnc$AREA)
# N/T Mean SD Min Max
# Overall 200 0.1263 0.0491 0.042 0.241
# Between 100 0.1263 0.0492 0.042 0.241
# Within 2 0.1263 0 0.1263 0.1263
settransform(pnc, AREA_diff = fdiff(AREA))
psmat(pnc$AREA_diff) |> head()
# 2000 2001
# 1825 NA 0
# 1827 NA 0
# 1828 NA 0
# 1831 NA 0
# 1832 NA 0
# 1833 NA 0
pnc <- unindex(pnc)
Unique Values, Ordering, Splitting, Binding
Functions funique() and roworder[v]() ignore the 'geometry' column in determining the unique values / order of rows when applied to sf data frames. rsplit() can be used to (recursively) split an sf data frame into multiple chunks.
# Splitting by SID74
rsplit(nc, ~ SID74) |> head(2)
# $`0`
# Simple feature collection with 13 features and 13 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 NWBIR74 BIR79 SID79 NWBIR79
# 1 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 10 542 3 12
# 2 0.062 1.547 1834 1834 Camden 37029 37029 15 286 115 350 2 139
# 3 0.091 1.284 1835 1835 Gates 37073 37073 37 420 254 594 2 371
# geometry
# 1 MULTIPOLYGON (((-81.23989 3...
# 2 MULTIPOLYGON (((-76.00897 3...
# 3 MULTIPOLYGON (((-76.56251 3...
#
# $`1`
# Simple feature collection with 11 features and 13 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 NWBIR74 BIR79 SID79 NWBIR79
# 1 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 10 1364 0 19
# 2 0.070 2.968 1831 1831 Currituck 37053 37053 27 508 123 830 2 145
# 3 0.124 1.428 1837 1837 Stokes 37169 37169 85 1612 160 2038 5 176
# geometry
# 1 MULTIPOLYGON (((-81.47276 3...
# 2 MULTIPOLYGON (((-76.00897 3...
# 3 MULTIPOLYGON (((-80.02567 3...
The default in rsplit() for data frames is simplify = TRUE, which, for a single LHS variable, would just split the column-vector. This does not apply to sf data frames as the 'geometry' column is always selected as well.
# Only splitting Area
rsplit(nc, AREA ~ SID74) |> head(1)
# $`0`
# Simple feature collection with 13 features and 1 field
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# AREA geometry
# 1 0.061 MULTIPOLYGON (((-81.23989 3...
# 2 0.062 MULTIPOLYGON (((-76.00897 3...
# 3 0.091 MULTIPOLYGON (((-76.56251 3...
# For data frames the default simplify = TRUE drops the data frame structure
rsplit(qDF(nc), AREA ~ SID74) |> head(1)
# $`0`
# [1] 0.061 0.062 0.091 0.064 0.059 0.080 0.066 0.099 0.094 0.078 0.131 0.167 0.051
sf data frames can be combined using rowbind(), which, by default, preserves the attributes of the first object.
# Splitting by each row and recombining
nc_combined <- nc %>% rsplit(seq_row(.)) %>% rowbind()
identical(nc, nc_combined)
# [1] TRUE
Transformations
For transforming and computing columns, fmutate() and ftransform[v]() apply as to any other data frame.
fmutate(nc, gsum_AREA = fsum(AREA, SID74, TRA = "fill")) |> head()
# Simple feature collection with 6 features and 15 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -81.74107 ymin: 36.07282 xmax: -75.77316 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79 SID79
# 1 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 1 10 1364 0
# 2 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 0 10 542 3
# 3 0.143 1.630 1828 1828 Surry 37171 37171 86 3188 5 208 3616 6
# NWBIR79 geometry gsum_AREA
# 1 19 MULTIPOLYGON (((-81.47276 3... 0.914
# 2 12 MULTIPOLYGON (((-81.23989 3... 1.103
# 3 260 MULTIPOLYGON (((-80.45634 3... 1.380
# Same thing, more expensive
nc |> fgroup_by(SID74) |> fmutate(gsum_AREA = fsum(AREA)) |> fungroup() |> head()
# Simple feature collection with 6 features and 15 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -81.74107 ymin: 36.07282 xmax: -75.77316 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79 SID79
# 1 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 1 10 1364 0
# 2 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 0 10 542 3
# 3 0.143 1.630 1828 1828 Surry 37171 37171 86 3188 5 208 3616 6
# NWBIR79 geometry gsum_AREA
# 1 19 MULTIPOLYGON (((-81.47276 3... 0.914
# 2 12 MULTIPOLYGON (((-81.23989 3... 1.103
# 3 260 MULTIPOLYGON (((-80.45634 3... 1.380
Special attention to sf data frames is afforded by fcompute(), which can be used to compute new columns dropping existing ones - except for the geometry column and any columns selected through the keep argument.
fcompute(nc, scaled_AREA = fscale(AREA),
gsum_AREA = fsum(AREA, SID74, TRA = "fill"),
keep = .c(AREA, SID74))
# Simple feature collection with 100 features and 4 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# AREA SID74 scaled_AREA gsum_AREA geometry
# 1 0.114 1 -0.2491860 0.914 MULTIPOLYGON (((-81.47276 3...
# 2 0.061 0 -1.3264176 1.103 MULTIPOLYGON (((-81.23989 3...
# 3 0.143 5 0.3402426 1.380 MULTIPOLYGON (((-80.45634 3...
Conversion to and from sf
The quick converters qDF(), qDT(), and qTBL() can be used to efficiently convert sf data frames to standard data frames, data.table's or tibbles, and the result can be converted back to the original sf data frame using setAttrib(), copyAttrib() or copyMostAttrib().
library(data.table)
# Create a data.table on the fly to do an fast grouped rolling mean and back to sf
qDT(nc)[, list(roll_AREA = frollmean(AREA, 2), geometry), by = SID74] |> copyMostAttrib(nc)
# Simple feature collection with 100 features and 2 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# SID74 roll_AREA geometry
# 1 1 NA MULTIPOLYGON (((-81.47276 3...
# 2 1 0.092 MULTIPOLYGON (((-76.00897 3...
# 3 1 0.097 MULTIPOLYGON (((-80.02567 3...
The easiest way to strip a geometry column off an sf data frame is via the function atomic_elem(), which removes list-like columns and, by default, also the class attribute. For example, we can create a data.table without list column using
qDT(atomic_elem(nc)) |> head()
# AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79 SID79
# <num> <num> <num> <num> <char> <char> <num> <int> <num> <num> <num> <num> <num>
# 1: 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 1 10 1364 0
# 2: 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 0 10 542 3
# 3: 0.143 1.630 1828 1828 Surry 37171 37171 86 3188 5 208 3616 6
# 4: 0.070 2.968 1831 1831 Currituck 37053 37053 27 508 1 123 830 2
# 5: 0.153 2.206 1832 1832 Northampton 37131 37131 66 1421 9 1066 1606 3
# 6: 0.097 1.670 1833 1833 Hertford 37091 37091 46 1452 7 954 1838 5
# NWBIR79
# <num>
# 1: 19
# 2: 12
# 3: 260
# 4: 145
# 5: 1197
# 6: 1237
This is also handy for other functions such as join() and pivot(), which are class agnostic like all of collapse, but do not have any built-in logic to deal with the sf column.
# Use atomic_elem() to strip geometry off y in left join
identical(nc, join(nc, atomic_elem(nc), overid = 2))
# left join: nc[AREA, PERIMETER, CNTY_, CNTY_ID, NAME, FIPS, FIPSNO, CRESS_ID, BIR74, SID74, NWBIR74, BIR79, SID79, NWBIR79] 100/100 (100%) <m:m> y[AREA, PERIMETER, CNTY_, CNTY_ID, NAME, FIPS, FIPSNO, CRESS_ID, BIR74, SID74, NWBIR74, BIR79, SID79, NWBIR79] 100/100 (100%)
# [1] TRUE
# In pivot: presently need to specify what to do with geometry column
pivot(nc, c("CNTY_ID", "geometry")) |> head()
# Simple feature collection with 6 features and 3 fields
# Geometry type: MULTIPOLYGON
# Dimension: XY
# Bounding box: xmin: -81.74107 ymin: 36.07282 xmax: -75.77316 ymax: 36.58965
# Geodetic CRS: NAD27
# First 3 features:
# CNTY_ID geometry variable value
# 1 1825 MULTIPOLYGON (((-81.47276 3... AREA 0.114
# 2 1827 MULTIPOLYGON (((-81.23989 3... AREA 0.061
# 3 1828 MULTIPOLYGON (((-80.45634 3... AREA 0.143
# Or use
pivot(qDT(atomic_elem(nc)), "CNTY_ID") |> head()
# CNTY_ID variable value
# <num> <fctr> <char>
# 1: 1825 AREA 0.114
# 2: 1827 AREA 0.061
# 3: 1828 AREA 0.143
# 4: 1831 AREA 0.07
# 5: 1832 AREA 0.153
# 6: 1833 AREA 0.097
Support for units
Since v2.0.13, collapse explicitly supports/preserves units objects through dedicated methods that preserve the 'units' class wherever sensible.
nc_dist <- st_centroid(nc) |> st_distance()
nc_dist[1:3, 1:3]
# Units: [m]
# [,1] [,2] [,3]
# [1,] 0.00 34020.35 72728.02
# [2,] 34020.35 0.00 40259.55
# [3,] 72728.02 40259.55 0.00
fmean(nc_dist) |> head()
# Units: [m]
# [1] 250543.9 237040.0 217941.5 337016.5 250380.2 269604.6
fndistinct(nc_dist) |> head()
# [1] 100 100 100 100 100 100
Conclusion
collapse provides no deep integration with the sf ecosystem and cannot perform spatial operations, but offers sufficient features and flexibility to painlessly manipulate sf data frames at much greater speeds than dplyr.
This requires a bit of care by the user though to ensure that the returned sf objects are valid, especially following aggregation and subsetting.
Try the collapse package in your browser
Any scripts or data that you put into this service are public.
collapse documentation built on Nov. 3, 2024, 9:08 a.m.
This short vignette focuses on using collapse with the popular sf package by Edzer Pebesma. It shows that collapse supports easy manipulation of sf data frames, at computation speeds far above dplyr.
collapse v1.6.0 added internal support for sf data frames by having most essential functions (e.g., fselect/gv, fsubset/ss, fgroup_by, findex_by, qsu, descr, varying, funique, roworder, rsplit, fcompute, ...) internally handle the geometry column.
To demonstrate this, we can load a test dataset provided by sf:
library(collapse) library(sf) nc <- st_read(system.file("shape/nc.shp", package = "sf"), quiet = TRUE) options(sf_max_print = 3) nc # Simple feature collection with 100 features and 14 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79 SID79 # 1 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 1 10 1364 0 # 2 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 0 10 542 3 # 3 0.143 1.630 1828 1828 Surry 37171 37171 86 3188 5 208 3616 6 # NWBIR79 geometry # 1 19 MULTIPOLYGON (((-81.47276 3... # 2 12 MULTIPOLYGON (((-81.23989 3... # 3 260 MULTIPOLYGON (((-80.45634 3...
Summarising sf Data Frames
Computing summary statistics on sf data frames automatically excludes the 'geometry' column:
# Which columns have at least 2 non-missing distinct values varying(nc) # AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 # TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE # NWBIR74 BIR79 SID79 NWBIR79 # TRUE TRUE TRUE TRUE # Quick summary stats qsu(nc) # N Mean SD Min Max # AREA 100 0.1263 0.0492 0.042 0.241 # PERIMETER 100 1.673 0.4823 0.999 3.64 # CNTY_ 100 1985.96 106.5166 1825 2241 # CNTY_ID 100 1985.96 106.5166 1825 2241 # NAME 100 - - - - # FIPS 100 - - - - # FIPSNO 100 37100 58.023 37001 37199 # CRESS_ID 100 50.5 29.0115 1 100 # BIR74 100 3299.62 3848.1651 248 21588 # SID74 100 6.67 7.7812 0 44 # NWBIR74 100 1050.81 1432.9117 1 8027 # BIR79 100 4223.92 5179.4582 319 30757 # SID79 100 8.36 9.4319 0 57 # NWBIR79 100 1352.81 1975.9988 3 11631 # Detailed statistics description of each column descr(nc) # Dataset: nc, 14 Variables, N = 100 # ---------------------------------------------------------------------------------------------------- # AREA (numeric): # Statistics # N Ndist Mean SD Min Max Skew Kurt # 100 77 0.13 0.05 0.04 0.24 0.48 2.5 # Quantiles # 1% 5% 10% 25% 50% 75% 90% 95% 99% # 0.04 0.06 0.06 0.09 0.12 0.15 0.2 0.21 0.24 # ---------------------------------------------------------------------------------------------------- # PERIMETER (numeric): # Statistics # N Ndist Mean SD Min Max Skew Kurt # 100 96 1.67 0.48 1 3.64 1.48 5.95 # Quantiles # 1% 5% 10% 25% 50% 75% 90% 95% 99% # 1 1.09 1.19 1.32 1.61 1.86 2.2 2.72 3.2 # ---------------------------------------------------------------------------------------------------- # CNTY_ (numeric): # Statistics # N Ndist Mean SD Min Max Skew Kurt # 100 100 1985.96 106.52 1825 2241 0.26 2.32 # Quantiles # 1% 5% 10% 25% 50% 75% 90% 95% 99% # 1826.98 1832.95 1837.9 1902.25 1982 2067.25 2110 2156.3 2238.03 # ---------------------------------------------------------------------------------------------------- # CNTY_ID (numeric): # Statistics # N Ndist Mean SD Min Max Skew Kurt # 100 100 1985.96 106.52 1825 2241 0.26 2.32 # Quantiles # 1% 5% 10% 25% 50% 75% 90% 95% 99% # 1826.98 1832.95 1837.9 1902.25 1982 2067.25 2110 2156.3 2238.03 # ---------------------------------------------------------------------------------------------------- # NAME (character): # Statistics # N Ndist # 100 100 # Table # Freq Perc # Ashe 1 1 # Alleghany 1 1 # Surry 1 1 # Currituck 1 1 # Northampton 1 1 # Hertford 1 1 # Camden 1 1 # Gates 1 1 # Warren 1 1 # Stokes 1 1 # Caswell 1 1 # Rockingham 1 1 # Granville 1 1 # Person 1 1 # ... 86 Others 86 86 # # Summary of Table Frequencies # Min. 1st Qu. Median Mean 3rd Qu. Max. # 1 1 1 1 1 1 # ---------------------------------------------------------------------------------------------------- # FIPS (character): # Statistics # N Ndist # 100 100 # Table # Freq Perc # 37009 1 1 # 37005 1 1 # 37171 1 1 # 37053 1 1 # 37131 1 1 # 37091 1 1 # 37029 1 1 # 37073 1 1 # 37185 1 1 # 37169 1 1 # 37033 1 1 # 37157 1 1 # 37077 1 1 # 37145 1 1 # ... 86 Others 86 86 # # Summary of Table Frequencies # Min. 1st Qu. Median Mean 3rd Qu. Max. # 1 1 1 1 1 1 # ---------------------------------------------------------------------------------------------------- # FIPSNO (numeric): # Statistics # N Ndist Mean SD Min Max Skew Kurt # 100 100 37100 58.02 37001 37199 -0 1.8 # Quantiles # 1% 5% 10% 25% 50% 75% 90% 95% 99% # 37002.98 37010.9 37020.8 37050.5 37100 37149.5 37179.2 37189.1 37197.02 # ---------------------------------------------------------------------------------------------------- # CRESS_ID (integer): # Statistics # N Ndist Mean SD Min Max Skew Kurt # 100 100 50.5 29.01 1 100 0 1.8 # Quantiles # 1% 5% 10% 25% 50% 75% 90% 95% 99% # 1.99 5.95 10.9 25.75 50.5 75.25 90.1 95.05 99.01 # ---------------------------------------------------------------------------------------------------- # BIR74 (numeric): # Statistics # N Ndist Mean SD Min Max Skew Kurt # 100 100 3299.62 3848.17 248 21588 2.79 11.79 # Quantiles # 1% 5% 10% 25% 50% 75% 90% 95% 99% # 283.64 419.75 531.8 1077 2180.5 3936 6725.7 11193 20378.22 # ---------------------------------------------------------------------------------------------------- # SID74 (numeric): # Statistics # N Ndist Mean SD Min Max Skew Kurt # 100 23 6.67 7.78 0 44 2.44 10.28 # Quantiles # 1% 5% 10% 25% 50% 75% 90% 95% 99% # 0 0 0 2 4 8.25 15.1 18.25 38.06 # ---------------------------------------------------------------------------------------------------- # NWBIR74 (numeric): # Statistics # N Ndist Mean SD Min Max Skew Kurt # 100 93 1050.81 1432.91 1 8027 2.83 11.84 # Quantiles # 1% 5% 10% 25% 50% 75% 90% 95% 99% # 1 9.95 39.2 190 697.5 1168.5 2231.8 3942.9 7052.84 # ---------------------------------------------------------------------------------------------------- # BIR79 (numeric): # Statistics # N Ndist Mean SD Min Max Skew Kurt # 100 100 4223.92 5179.46 319 30757 2.99 13.1 # Quantiles # 1% 5% 10% 25% 50% 75% 90% 95% 99% # 349.69 539.3 675.7 1336.25 2636 4889 8313 14707.45 26413.87 # ---------------------------------------------------------------------------------------------------- # SID79 (numeric): # Statistics # N Ndist Mean SD Min Max Skew Kurt # 100 28 8.36 9.43 0 57 2.28 9.88 # Quantiles # 1% 5% 10% 25% 50% 75% 90% 95% 99% # 0 0 1 2 5 10.25 21 26 38.19 # ---------------------------------------------------------------------------------------------------- # NWBIR79 (numeric): # Statistics # N Ndist Mean SD Min Max Skew Kurt # 100 98 1352.81 1976 3 11631 3.18 14.45 # Quantiles # 1% 5% 10% 25% 50% 75% 90% 95% 99% # 3.99 11.9 44.7 250.5 874.5 1406.75 2987.9 5090.5 10624.17 # ----------------------------------------------------------------------------------------------------
Selecting Columns and Subsetting
We can select columns from the sf data frame without having to worry about taking along 'geometry':
# Selecting a sequence of columns fselect(nc, AREA, NAME:FIPSNO) # Simple feature collection with 100 features and 4 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # AREA NAME FIPS FIPSNO geometry # 1 0.114 Ashe 37009 37009 MULTIPOLYGON (((-81.47276 3... # 2 0.061 Alleghany 37005 37005 MULTIPOLYGON (((-81.23989 3... # 3 0.143 Surry 37171 37171 MULTIPOLYGON (((-80.45634 3... # Same using standard evaluation (gv is a shorthand for get_vars()) gv(nc, c("AREA", "NAME", "FIPS", "FIPSNO")) # Simple feature collection with 100 features and 4 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # AREA NAME FIPS FIPSNO geometry # 1 0.114 Ashe 37009 37009 MULTIPOLYGON (((-81.47276 3... # 2 0.061 Alleghany 37005 37005 MULTIPOLYGON (((-81.23989 3... # 3 0.143 Surry 37171 37171 MULTIPOLYGON (((-80.45634 3...
The same applies to subsetting rows (and columns):
# A fast and enhanced version of base::subset fsubset(nc, AREA > fmean(AREA), AREA, NAME:FIPSNO) # Simple feature collection with 44 features and 4 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # AREA NAME FIPS FIPSNO geometry # 1 0.143 Surry 37171 37171 MULTIPOLYGON (((-80.45634 3... # 2 0.153 Northampton 37131 37131 MULTIPOLYGON (((-77.21767 3... # 3 0.153 Rockingham 37157 37157 MULTIPOLYGON (((-79.53051 3... # A fast version of `[` (where i is used and optionally j) ss(nc, 1:10, c("AREA", "NAME", "FIPS", "FIPSNO")) # Simple feature collection with 10 features and 4 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # AREA NAME FIPS FIPSNO geometry # 1 0.114 Ashe 37009 37009 MULTIPOLYGON (((-81.47276 3... # 2 0.061 Alleghany 37005 37005 MULTIPOLYGON (((-81.23989 3... # 3 0.143 Surry 37171 37171 MULTIPOLYGON (((-80.45634 3...
This is significantly faster than using [, base::subset(), dplyr::select() or dplyr::filter():
library(microbenchmark) library(dplyr) # Selecting columns microbenchmark(collapse = fselect(nc, AREA, NAME:FIPSNO), dplyr = select(nc, AREA, NAME:FIPSNO), collapse2 = gv(nc, c("AREA", "NAME", "FIPS", "FIPSNO")), sf = nc[c("AREA", "NAME", "FIPS", "FIPSNO")]) # Unit: microseconds # expr min lq mean median uq max neval # collapse 3.034 3.9565 5.19429 5.1865 5.6990 22.878 100 # dplyr 431.279 452.2915 505.29015 466.3750 493.8450 3356.342 100 # collapse2 2.665 3.4850 4.59610 4.4075 5.0635 14.391 100 # sf 105.165 114.1235 120.39732 118.0390 124.9270 156.497 100 # Subsetting microbenchmark(collapse = fsubset(nc, AREA > fmean(AREA), AREA, NAME:FIPSNO), dplyr = select(nc, AREA, NAME:FIPSNO) |> filter(AREA > fmean(AREA)), collapse2 = ss(nc, 1:10, c("AREA", "NAME", "FIPS", "FIPSNO")), sf = nc[1:10, c("AREA", "NAME", "FIPS", "FIPSNO")]) # Unit: microseconds # expr min lq mean median uq max neval # collapse 9.676 11.5825 15.01707 14.4730 16.8920 30.463 100 # dplyr 890.643 917.6415 1055.40970 941.7085 1009.7890 5546.685 100 # collapse2 2.829 3.5465 5.40585 4.8995 6.4165 20.541 100 # sf 176.997 187.6160 202.72286 200.7565 210.8220 340.464 100
However, collapse functions don't subset the 'agr' attribute on selecting columns, which (if specified) relates columns (attributes) to the geometry, and also don't modify the 'bbox' attribute giving the overall boundaries of a set of geometries when subsetting the sf data frame. Keeping the full 'agr' attribute is not problematic for all practical purposes, but not changing 'bbox' upon subsetting may lead to too large margins when plotting the geometries of a subset sf data frame.
One way to to change this is calling st_make_valid() on the subset frame; but st_make_valid() is very expensive, thus unless the subset frame is very small, it is better to use [, base::subset() or dplyr::filter() in cases where the bounding box size matters.
Aggregation and Grouping
The flexibility and speed of collap() for aggregation can be used on sf data frames. A separate method for sf objects was not considered necessary as one can simply aggregate the geometry column using st_union():
# Aggregating by variable SID74 using the median for numeric and the mode for categorical columns collap(nc, ~ SID74, custom = list(fmedian = is.numeric, fmode = is.character, st_union = "geometry")) # or use is.list to fetch the geometry # Simple feature collection with 23 features and 15 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 SID74 NWBIR74 BIR79 # 1 0.0780 1.3070 1950.0 1950.0 Alleghany 37005 37073 37.0 487 0 0 40.0 594.0 # 2 0.0810 1.2880 1887.0 1887.0 Ashe 37009 37137 69.0 751 1 1 148.0 899.0 # 3 0.1225 1.6435 1959.5 1959.5 Caswell 37033 37078 39.5 1271 2 2 382.5 1676.5 # SID79 NWBIR79 geometry # 1 1 45 MULTIPOLYGON (((-83.69563 3... # 2 1 176 MULTIPOLYGON (((-80.02406 3... # 3 2 452 MULTIPOLYGON (((-77.16129 3...
sf data frames can also be grouped and then aggregated using fsummarise():
nc |> fgroup_by(SID74) # Simple feature collection with 100 features and 14 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79 SID79 # 1 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 1 10 1364 0 # 2 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 0 10 542 3 # 3 0.143 1.630 1828 1828 Surry 37171 37171 86 3188 5 208 3616 6 # NWBIR79 geometry # 1 19 MULTIPOLYGON (((-81.47276 3... # 2 12 MULTIPOLYGON (((-81.23989 3... # 3 260 MULTIPOLYGON (((-80.45634 3... # # Grouped by: SID74 [23 | 4 (4) 1-13] nc |> fgroup_by(SID74) |> fsummarise(AREA_Ag = fsum(AREA), Perimeter_Ag = fmedian(PERIMETER), geometry = st_union(geometry)) # Simple feature collection with 23 features and 3 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # SID74 AREA_Ag Perimeter_Ag geometry # 1 0 1.103 1.3070 MULTIPOLYGON (((-83.69563 3... # 2 1 0.914 1.2880 MULTIPOLYGON (((-80.02406 3... # 3 2 1.047 1.6435 MULTIPOLYGON (((-77.16129 3...
Typically most of the time in aggregation is consumed by st_union() so that the speed of collapse does not really become visible on most datasets. A faster alternative is to use geos (sf backend for planar geometries) or s2 (sf backend for spherical geometries) directly:
# Using s2 backend: sensible for larger tasks nc |> fmutate(geometry = s2::as_s2_geography(geometry)) |> fgroup_by(SID74) |> fsummarise(AREA_Ag = fsum(AREA), Perimeter_Ag = fmedian(PERIMETER), geometry = s2::s2_union_agg(geometry)) |> fmutate(geometry = st_as_sfc(geometry)) # Simple feature collection with 23 features and 3 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: WGS 84 # First 3 features: # SID74 AREA_Ag Perimeter_Ag geometry # 1 0 1.103 1.3070 MULTIPOLYGON (((-83.69563 3... # 2 1 0.914 1.2880 MULTIPOLYGON (((-80.02406 3... # 3 2 1.047 1.6435 MULTIPOLYGON (((-77.16129 3...
In general, also upon aggregation with collapse, functions st_as_sfc(), st_as_sf(), or, in the worst case, st_make_valid(), may need to be invoked to ensure valid sf object output. Functions collap() and fsummarise() are attribute preserving but do not give special regard to geometry columns.
One exception that both avoids the high cost of spatial functions in aggregation and any need for ex-post conversion/validation is aggregating spatial panel data over the time-dimension. Such panels can quickly be aggregated using ffirst() or flast() to aggregate the geometry:
# Creating a panel-dataset by simply duplicating nc for 2 different years pnc <- rowbind(`2000` = nc, `2001` = nc, idcol = "Year") |> as_integer_factor() pnc # Simple feature collection with 200 features and 15 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # Year AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79 # 1 2000 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 1 10 1364 # 2 2000 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 0 10 542 # 3 2000 0.143 1.630 1828 1828 Surry 37171 37171 86 3188 5 208 3616 # SID79 NWBIR79 geometry # 1 0 19 MULTIPOLYGON (((-81.47276 3... # 2 3 12 MULTIPOLYGON (((-81.23989 3... # 3 6 260 MULTIPOLYGON (((-80.45634 3... # Aggregating by NAME, using the last value for all categorical data collap(pnc, ~ NAME, fmedian, catFUN = flast, cols = -1L) # Simple feature collection with 100 features and 15 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # AREA PERIMETER CNTY_ CNTY_ID NAME NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79 # 1 0.111 1.392 1904 1904 Alamance Alamance 37001 37001 1 4672 13 1243 5767 # 2 0.066 1.070 1950 1950 Alexander Alexander 37003 37003 2 1333 0 128 1683 # 3 0.061 1.231 1827 1827 Alleghany Alleghany 37005 37005 3 487 0 10 542 # SID79 NWBIR79 geometry # 1 11 1397 MULTIPOLYGON (((-79.24619 3... # 2 2 150 MULTIPOLYGON (((-81.10889 3... # 3 3 12 MULTIPOLYGON (((-81.23989 3... # Using fsummarise to aggregate just two variables and the geometry pnc_ag <- pnc |> fgroup_by(NAME) |> fsummarise(AREA_Ag = fsum(AREA), Perimeter_Ag = fmedian(PERIMETER), geometry = flast(geometry)) # The geometry is still valid... (slt = shorthand for fselect) plot(slt(pnc_ag, AREA_Ag))

Indexing
sf data frames can also become indexed frames (spatio-temporal panels):
pnc <- pnc |> findex_by(CNTY_ID, Year) pnc # Simple feature collection with 200 features and 15 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # Year AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79 # 1 2000 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 1 10 1364 # 2 2000 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 0 10 542 # 3 2000 0.143 1.630 1828 1828 Surry 37171 37171 86 3188 5 208 3616 # SID79 NWBIR79 geometry # 1 0 19 MULTIPOLYGON (((-81.47276 3... # 2 3 12 MULTIPOLYGON (((-81.23989 3... # 3 6 260 MULTIPOLYGON (((-80.45634 3... # # Indexed by: CNTY_ID [100] | Year [2] qsu(pnc$AREA) # N/T Mean SD Min Max # Overall 200 0.1263 0.0491 0.042 0.241 # Between 100 0.1263 0.0492 0.042 0.241 # Within 2 0.1263 0 0.1263 0.1263 settransform(pnc, AREA_diff = fdiff(AREA)) psmat(pnc$AREA_diff) |> head() # 2000 2001 # 1825 NA 0 # 1827 NA 0 # 1828 NA 0 # 1831 NA 0 # 1832 NA 0 # 1833 NA 0 pnc <- unindex(pnc)
Unique Values, Ordering, Splitting, Binding
Functions funique() and roworder[v]() ignore the 'geometry' column in determining the unique values / order of rows when applied to sf data frames. rsplit() can be used to (recursively) split an sf data frame into multiple chunks.
# Splitting by SID74 rsplit(nc, ~ SID74) |> head(2) # $`0` # Simple feature collection with 13 features and 13 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 NWBIR74 BIR79 SID79 NWBIR79 # 1 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 10 542 3 12 # 2 0.062 1.547 1834 1834 Camden 37029 37029 15 286 115 350 2 139 # 3 0.091 1.284 1835 1835 Gates 37073 37073 37 420 254 594 2 371 # geometry # 1 MULTIPOLYGON (((-81.23989 3... # 2 MULTIPOLYGON (((-76.00897 3... # 3 MULTIPOLYGON (((-76.56251 3... # # $`1` # Simple feature collection with 11 features and 13 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 NWBIR74 BIR79 SID79 NWBIR79 # 1 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 10 1364 0 19 # 2 0.070 2.968 1831 1831 Currituck 37053 37053 27 508 123 830 2 145 # 3 0.124 1.428 1837 1837 Stokes 37169 37169 85 1612 160 2038 5 176 # geometry # 1 MULTIPOLYGON (((-81.47276 3... # 2 MULTIPOLYGON (((-76.00897 3... # 3 MULTIPOLYGON (((-80.02567 3...
The default in rsplit() for data frames is simplify = TRUE, which, for a single LHS variable, would just split the column-vector. This does not apply to sf data frames as the 'geometry' column is always selected as well.
# Only splitting Area rsplit(nc, AREA ~ SID74) |> head(1) # $`0` # Simple feature collection with 13 features and 1 field # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # AREA geometry # 1 0.061 MULTIPOLYGON (((-81.23989 3... # 2 0.062 MULTIPOLYGON (((-76.00897 3... # 3 0.091 MULTIPOLYGON (((-76.56251 3... # For data frames the default simplify = TRUE drops the data frame structure rsplit(qDF(nc), AREA ~ SID74) |> head(1) # $`0` # [1] 0.061 0.062 0.091 0.064 0.059 0.080 0.066 0.099 0.094 0.078 0.131 0.167 0.051
sf data frames can be combined using rowbind(), which, by default, preserves the attributes of the first object.
# Splitting by each row and recombining nc_combined <- nc %>% rsplit(seq_row(.)) %>% rowbind() identical(nc, nc_combined) # [1] TRUE
Transformations
For transforming and computing columns, fmutate() and ftransform[v]() apply as to any other data frame.
fmutate(nc, gsum_AREA = fsum(AREA, SID74, TRA = "fill")) |> head() # Simple feature collection with 6 features and 15 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -81.74107 ymin: 36.07282 xmax: -75.77316 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79 SID79 # 1 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 1 10 1364 0 # 2 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 0 10 542 3 # 3 0.143 1.630 1828 1828 Surry 37171 37171 86 3188 5 208 3616 6 # NWBIR79 geometry gsum_AREA # 1 19 MULTIPOLYGON (((-81.47276 3... 0.914 # 2 12 MULTIPOLYGON (((-81.23989 3... 1.103 # 3 260 MULTIPOLYGON (((-80.45634 3... 1.380 # Same thing, more expensive nc |> fgroup_by(SID74) |> fmutate(gsum_AREA = fsum(AREA)) |> fungroup() |> head() # Simple feature collection with 6 features and 15 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -81.74107 ymin: 36.07282 xmax: -75.77316 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79 SID79 # 1 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 1 10 1364 0 # 2 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 0 10 542 3 # 3 0.143 1.630 1828 1828 Surry 37171 37171 86 3188 5 208 3616 6 # NWBIR79 geometry gsum_AREA # 1 19 MULTIPOLYGON (((-81.47276 3... 0.914 # 2 12 MULTIPOLYGON (((-81.23989 3... 1.103 # 3 260 MULTIPOLYGON (((-80.45634 3... 1.380
Special attention to sf data frames is afforded by fcompute(), which can be used to compute new columns dropping existing ones - except for the geometry column and any columns selected through the keep argument.
fcompute(nc, scaled_AREA = fscale(AREA), gsum_AREA = fsum(AREA, SID74, TRA = "fill"), keep = .c(AREA, SID74)) # Simple feature collection with 100 features and 4 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # AREA SID74 scaled_AREA gsum_AREA geometry # 1 0.114 1 -0.2491860 0.914 MULTIPOLYGON (((-81.47276 3... # 2 0.061 0 -1.3264176 1.103 MULTIPOLYGON (((-81.23989 3... # 3 0.143 5 0.3402426 1.380 MULTIPOLYGON (((-80.45634 3...
Conversion to and from sf
The quick converters qDF(), qDT(), and qTBL() can be used to efficiently convert sf data frames to standard data frames, data.table's or tibbles, and the result can be converted back to the original sf data frame using setAttrib(), copyAttrib() or copyMostAttrib().
library(data.table) # Create a data.table on the fly to do an fast grouped rolling mean and back to sf qDT(nc)[, list(roll_AREA = frollmean(AREA, 2), geometry), by = SID74] |> copyMostAttrib(nc) # Simple feature collection with 100 features and 2 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # SID74 roll_AREA geometry # 1 1 NA MULTIPOLYGON (((-81.47276 3... # 2 1 0.092 MULTIPOLYGON (((-76.00897 3... # 3 1 0.097 MULTIPOLYGON (((-80.02567 3...
The easiest way to strip a geometry column off an sf data frame is via the function atomic_elem(), which removes list-like columns and, by default, also the class attribute. For example, we can create a data.table without list column using
qDT(atomic_elem(nc)) |> head() # AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74 SID74 NWBIR74 BIR79 SID79 # <num> <num> <num> <num> <char> <char> <num> <int> <num> <num> <num> <num> <num> # 1: 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091 1 10 1364 0 # 2: 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487 0 10 542 3 # 3: 0.143 1.630 1828 1828 Surry 37171 37171 86 3188 5 208 3616 6 # 4: 0.070 2.968 1831 1831 Currituck 37053 37053 27 508 1 123 830 2 # 5: 0.153 2.206 1832 1832 Northampton 37131 37131 66 1421 9 1066 1606 3 # 6: 0.097 1.670 1833 1833 Hertford 37091 37091 46 1452 7 954 1838 5 # NWBIR79 # <num> # 1: 19 # 2: 12 # 3: 260 # 4: 145 # 5: 1197 # 6: 1237
This is also handy for other functions such as join() and pivot(), which are class agnostic like all of collapse, but do not have any built-in logic to deal with the sf column.
# Use atomic_elem() to strip geometry off y in left join identical(nc, join(nc, atomic_elem(nc), overid = 2)) # left join: nc[AREA, PERIMETER, CNTY_, CNTY_ID, NAME, FIPS, FIPSNO, CRESS_ID, BIR74, SID74, NWBIR74, BIR79, SID79, NWBIR79] 100/100 (100%) <m:m> y[AREA, PERIMETER, CNTY_, CNTY_ID, NAME, FIPS, FIPSNO, CRESS_ID, BIR74, SID74, NWBIR74, BIR79, SID79, NWBIR79] 100/100 (100%) # [1] TRUE # In pivot: presently need to specify what to do with geometry column pivot(nc, c("CNTY_ID", "geometry")) |> head() # Simple feature collection with 6 features and 3 fields # Geometry type: MULTIPOLYGON # Dimension: XY # Bounding box: xmin: -81.74107 ymin: 36.07282 xmax: -75.77316 ymax: 36.58965 # Geodetic CRS: NAD27 # First 3 features: # CNTY_ID geometry variable value # 1 1825 MULTIPOLYGON (((-81.47276 3... AREA 0.114 # 2 1827 MULTIPOLYGON (((-81.23989 3... AREA 0.061 # 3 1828 MULTIPOLYGON (((-80.45634 3... AREA 0.143 # Or use pivot(qDT(atomic_elem(nc)), "CNTY_ID") |> head() # CNTY_ID variable value # <num> <fctr> <char> # 1: 1825 AREA 0.114 # 2: 1827 AREA 0.061 # 3: 1828 AREA 0.143 # 4: 1831 AREA 0.07 # 5: 1832 AREA 0.153 # 6: 1833 AREA 0.097
Support for units
Since v2.0.13, collapse explicitly supports/preserves units objects through dedicated methods that preserve the 'units' class wherever sensible.
nc_dist <- st_centroid(nc) |> st_distance() nc_dist[1:3, 1:3] # Units: [m] # [,1] [,2] [,3] # [1,] 0.00 34020.35 72728.02 # [2,] 34020.35 0.00 40259.55 # [3,] 72728.02 40259.55 0.00 fmean(nc_dist) |> head() # Units: [m] # [1] 250543.9 237040.0 217941.5 337016.5 250380.2 269604.6 fndistinct(nc_dist) |> head() # [1] 100 100 100 100 100 100
Conclusion
collapse provides no deep integration with the sf ecosystem and cannot perform spatial operations, but offers sufficient features and flexibility to painlessly manipulate sf data frames at much greater speeds than dplyr.
This requires a bit of care by the user though to ensure that the returned sf objects are valid, especially following aggregation and subsetting.
Try the collapse package in your browser
Any scripts or data that you put into this service are public.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
