Getting started with the `makeit` package
In makeit: Run R Scripts if Needed
knitr::opts_chunk$set(collapse=TRUE, comment="#>")
library(makeit)
unlink("examples", recursive=TRUE)
file.copy(system.file("examples", package="makeit"), ".", recursive=TRUE)
```{css, echo=FALSE}
div.sourceCode {margin-bottom:3ex}
h1 {font-size:3ex; margin-top:3ex}
h2 {margin-top:2ex}
img {margin-bottom:1ex; margin-top:1ex}
p {margin-top:2ex}
pre {margin-bottom:3ex; margin-top:2ex}
table {margin-bottom:3ex}
# Overview
The [makeit](https://cran.r-project.org/package=makeit) package provides a
simple [make](https://en.wikipedia.org/wiki/Make_(software))-like utility to run
R scripts if needed, based on the last modified time. It comes with no package
dependencies, organizational overhead, or structural requirements.
The general idea is to run a workflow without repeating tasks that are already
completed. A workflow consists of one or more R scripts, where each script
generates output files.
# Tutorials
The following tutorials come with the package and can be copied from
`library/makeit/examples` to a working directory, or downloaded from
[GitHub](https://github.com/arni-magnusson/makeit/tree/main/inst/examples).
## analysis
```r
knitr::opts_knit$set(root.dir="examples/analysis")
This example consists of a script analysis.R that uses input.dat to produce
output.dat.
Before
cat(dir(), sep="\n")
Run
make("analysis.R", "input.dat", "output.dat")
Try running again:
make("analysis.R", "input.dat", "output.dat")
Note how a make() call has the general form: script x uses y to produce
z.
After
cat(dir(), sep="\n")
sequential
knitr::opts_knit$set(root.dir="examples/sequential")
This example consists of three scripts, where one runs after the other.
The plot script produces files inside a plots folder and the table script
produces files inside a tables folder.
Before
cat(dir()[dir() != "_make.R"], sep="\n")
Run
make("01_model.R", "data.dat", "results.dat")
make("02_plots.R", "results.dat", c("plots/A.png", "plots/B.png"))
make("03_tables.R", "results.dat", c("tables/A.csv", "tables/B.csv"))
For convenience, a _make.R file is provided, containing these make() calls.
After
files <- dir(recursive=TRUE)
files <- files[files != "_make.R"]
files <- c(grep("/", files, value=TRUE),
grep("/", files, value=TRUE, invert=TRUE))
cat(files, sep="\n")
four_minutes
knitr::opts_knit$set(root.dir="examples/four_minutes")
Similar to the 'sequential' example above, but based on the
four-minutes tutorial that
comes with targets package.
Before
cat(dir()[dir() != "_make.R"], sep="\n")
Run
make("get_data.R", "data_raw.csv", "data/data.csv")
make("fit_model.R", "data/data.csv", "output/coefs.dat")
make("plot_model.R", c("data/data.csv", "output/coefs.dat"), "output/plot.pdf")
For convenience, a _make.R file is provided, containing these make() calls.
After
files <- dir(recursive=TRUE)
files <- files[files != "_make.R"]
files <- c(grep("/", files, value=TRUE),
grep("/", files, value=TRUE, invert=TRUE))
cat(files, sep="\n")
dag_wikipedia
knitr::opts_knit$set(root.dir="examples/dag_wikipedia")
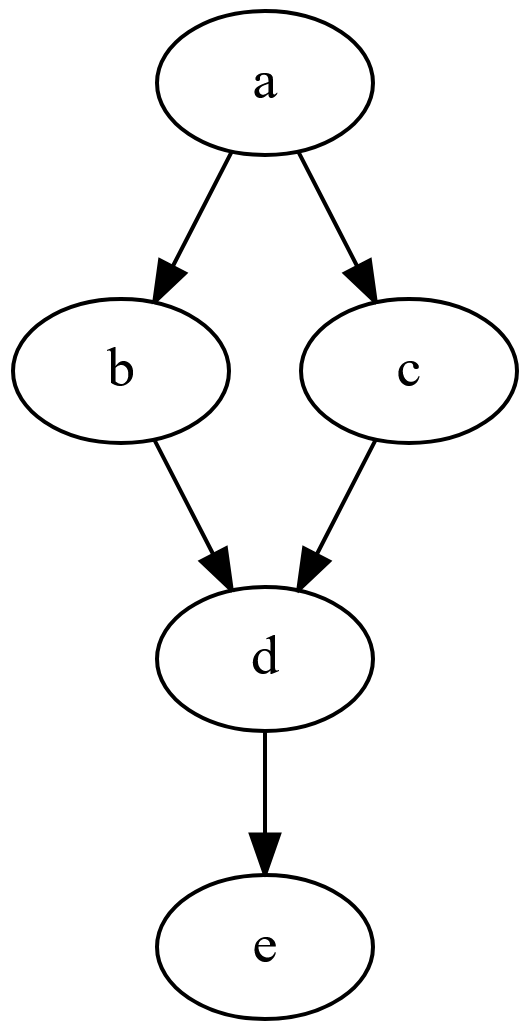
DAG example based on the diagram provided in the Wikipedia article on directed
acyclic
graph.
Each script produces a corresponding output file: a.R produces out/a.dat,
b.R produces out/b.dat, etc.
Before
cat(dir()[dir() != "_make.R"], sep="\n")
Run
make("a.R", prereq=NULL, target="out/a.dat")
make("b.R", prereq="out/a.dat", target="out/b.dat")
make("c.R", prereq="out/a.dat", target="out/c.dat")
make("d.R", prereq=c("out/b.dat", "out/c.dat"), target="out/d.dat")
make("e.R", prereq="out/d.dat", target="out/e.dat")
For convenience, a _make.R file is provided, containing these make() calls.
After
files <- dir(recursive=TRUE)
files <- files[files != "_make.R"]
files <- c(grep("/", files, value=TRUE),
grep("/", files, value=TRUE, invert=TRUE))
cat(files, sep="\n")
dag_targets
knitr::opts_knit$set(root.dir="examples/dag_targets")
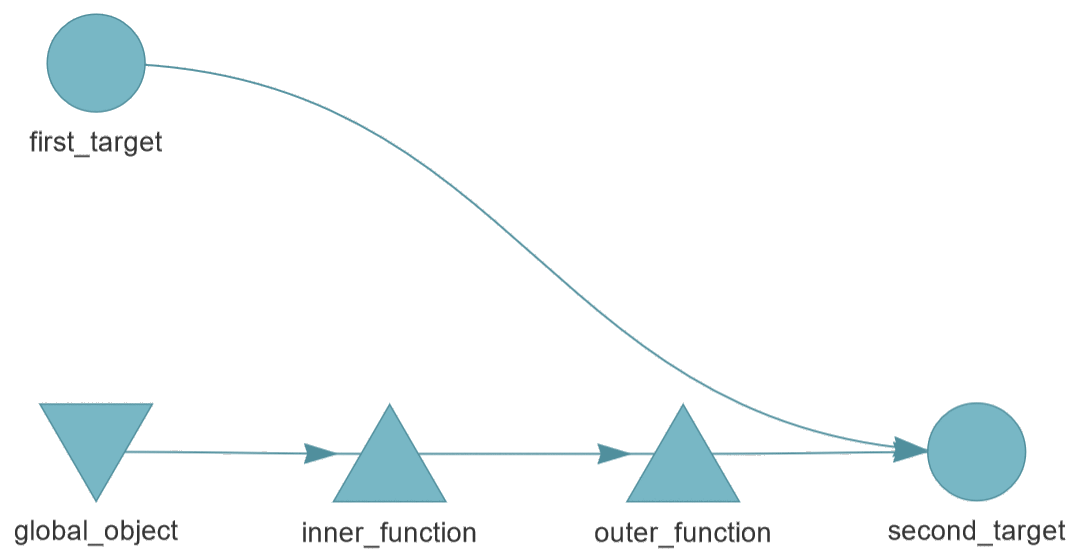
DAG example based on the example from the targets user
manual.
The second_target depends on first_target and outer_function, which in
turn depends on inner_function and global_object.
Before
cat(dir()[dir() != "_make.R"], sep="\n")
Run
make("first_target.R", NULL, "output/first_target.dat")
make("global_object.R", NULL, "output/global_object.dat")
make("second_target.R",
prereq=c("output/first_target.dat", "output/global_object.dat",
"inner_function.R", "outer_function.R"),
target="output/second_target.dat")
For convenience, a _make.R file is provided, containing these make() calls.
After
files <- dir(recursive=TRUE)
files <- files[files != "_make.R"]
files <- c(grep("/", files, value=TRUE),
grep("/", files, value=TRUE, invert=TRUE))
cat(files, sep="\n")
Discussion
Use cases
The make() function is a tool that can be applied to many types of workflows,
consisting of one or many R scripts. It is especially useful when the complete
workflow takes many minutes or hours to run. Changing one part of the analysis
will then update the related plots and tables, without rerunning every part of
the analysis.
Your project
Most analyses resemble the sequential example above, dividing the workflow
into steps that run one after another. As an introductory example, the
sequential workflow consists of only three steps: model, plots, and tables.
In practice, it is usually practical to divide a workflow into more steps than
that, the first step being data preparation, such as importing, filtering,
aggregating, and converting the data to the format that the model expects.
If the model is non-trivial, it can be practical to have an output.R step
righter after model.R, extracting the results from the model-specific format to
a more general format that is easy to browse, tabulate, and plot. This way, the
model.R script can be very short, making it easy to see and understand the
modelling approach and configuration. Separating the fundamental modelling step
from the manual labor of data preparation and plotting can make an analysis more
open and reproducible - for others to browse and reuse.
The paradigm of using small dedicated scripts with clear input and output files
(read and write function calls near the beginning and end of each script) is
usually a better workflow design than managing a large monolithic script where
the user navigates between sections to run selected blocks of code.
Comparison with other packages
The four_minutes and dag_targets examples above provide an interesting
comparison between the makeit package and the targets package, for example.
makeit
The makeit package is script-based, where each step passes the results to the
next step as output files. The user organizes their workflow by writing scripts
that produce files.
The makeit package has no package dependencies, takes a very short time to
learn, and can be used to run any existing workflows, as long as they are based
on scripts with input and output files. The scripts may include functions, but
that is not a requirement.
The package consists of a single function that does one thing: run an R script
if underlying files have changed, otherwise do nothing.
TAF
The TAF package contains a similar make() function and is an ancestor of the
makeit package. The overall aim of TAF is to support and strengthen
reproducibility in science, as well as reviewability.
The TAF package provides a structured modular design that divides a workflow
into four main stages: data.R, model.R, output.R, and report.R. The
initial data are declared in a DATA.bib file and an optional SOFTWARE.bib
file can be used to declare specific versions of R packages and other software.
The package consists of many useful tools to support reproducible workflows for
scientific analyses.
targets
The targets package is function-based, where each step passes the results to
the next step as objects in memory. The user organizes their workflow by writing
functions that produce objects. It is the successor of the older drake
package.
The targets package relies on many underlying packages, takes some time to
learn, and some work may be required to realign existing workflows into
functions. The functions may produce files, but that is not a requirement.
The package consists of many useful tools to support workflow design and
management.
Comparison
Package | Paradigm | State | Package dependencies | Time to learn | Run existing workflow | Features
--------- | --------- | ------ | -------------------- | ------------- | ---------------------- | --------
makeit | Scripts | Files | None | Very short | Must be file-based | One
TAF | Scripts | Files | None | Some | Must be file-based | Many
targets | Functions | Memory | Many | Some | Must be function-based | Many
The CRAN task view for reproducible
research provides an
annotated list of packages related to pipeline
toolkits
and project
workflows.
References
Magnusson, A. makeit: Run R Scripts if Needed.\
https://cran.r-project.org/package=makeit
Magnusson, A. and C. Millar. TAF: Transparent Assessment Framework for
Reproducible Research.\
https://cran.r-project.org/package=TAF
Landau, W.M. 2021. The targets R package: a dynamic Make-like function-oriented
pipeline toolkit for reproducibility and high-performance computing. Journal of
Open Source Software, 6(57), 2959.\
https://doi.org/10.21105/joss.02959, https://cran.r-project.org/package=targets
Stallman, R.M. et al. An introduction to makefiles. Chapter 2 in the GNU Make
manual.\
https://www.gnu.org/software/make/manual/
Try the makeit package in your browser
Any scripts or data that you put into this service are public.
makeit documentation built on April 11, 2025, 5:38 p.m.
knitr::opts_chunk$set(collapse=TRUE, comment="#>") library(makeit) unlink("examples", recursive=TRUE) file.copy(system.file("examples", package="makeit"), ".", recursive=TRUE)
```{css, echo=FALSE} div.sourceCode {margin-bottom:3ex} h1 {font-size:3ex; margin-top:3ex} h2 {margin-top:2ex} img {margin-bottom:1ex; margin-top:1ex} p {margin-top:2ex} pre {margin-bottom:3ex; margin-top:2ex} table {margin-bottom:3ex}
# Overview The [makeit](https://cran.r-project.org/package=makeit) package provides a simple [make](https://en.wikipedia.org/wiki/Make_(software))-like utility to run R scripts if needed, based on the last modified time. It comes with no package dependencies, organizational overhead, or structural requirements. The general idea is to run a workflow without repeating tasks that are already completed. A workflow consists of one or more R scripts, where each script generates output files. # Tutorials The following tutorials come with the package and can be copied from `library/makeit/examples` to a working directory, or downloaded from [GitHub](https://github.com/arni-magnusson/makeit/tree/main/inst/examples). ## analysis ```r knitr::opts_knit$set(root.dir="examples/analysis")
This example consists of a script analysis.R that uses input.dat to produce
output.dat.
Before
cat(dir(), sep="\n")
Run
make("analysis.R", "input.dat", "output.dat")
Try running again:
make("analysis.R", "input.dat", "output.dat")
Note how a make() call has the general form: script x uses y to produce
z.
After
cat(dir(), sep="\n")
sequential
knitr::opts_knit$set(root.dir="examples/sequential")
This example consists of three scripts, where one runs after the other.
The plot script produces files inside a plots folder and the table script
produces files inside a tables folder.
Before
cat(dir()[dir() != "_make.R"], sep="\n")
Run
make("01_model.R", "data.dat", "results.dat") make("02_plots.R", "results.dat", c("plots/A.png", "plots/B.png")) make("03_tables.R", "results.dat", c("tables/A.csv", "tables/B.csv"))
For convenience, a _make.R file is provided, containing these make() calls.
After
files <- dir(recursive=TRUE) files <- files[files != "_make.R"] files <- c(grep("/", files, value=TRUE), grep("/", files, value=TRUE, invert=TRUE)) cat(files, sep="\n")
four_minutes
knitr::opts_knit$set(root.dir="examples/four_minutes")
Similar to the 'sequential' example above, but based on the
four-minutes tutorial that
comes with targets package.
Before
cat(dir()[dir() != "_make.R"], sep="\n")
Run
make("get_data.R", "data_raw.csv", "data/data.csv") make("fit_model.R", "data/data.csv", "output/coefs.dat") make("plot_model.R", c("data/data.csv", "output/coefs.dat"), "output/plot.pdf")
For convenience, a _make.R file is provided, containing these make() calls.
After
files <- dir(recursive=TRUE) files <- files[files != "_make.R"] files <- c(grep("/", files, value=TRUE), grep("/", files, value=TRUE, invert=TRUE)) cat(files, sep="\n")
dag_wikipedia
knitr::opts_knit$set(root.dir="examples/dag_wikipedia")

DAG example based on the diagram provided in the Wikipedia article on directed acyclic graph.
Each script produces a corresponding output file: a.R produces out/a.dat,
b.R produces out/b.dat, etc.
Before
cat(dir()[dir() != "_make.R"], sep="\n")
Run
make("a.R", prereq=NULL, target="out/a.dat") make("b.R", prereq="out/a.dat", target="out/b.dat") make("c.R", prereq="out/a.dat", target="out/c.dat") make("d.R", prereq=c("out/b.dat", "out/c.dat"), target="out/d.dat") make("e.R", prereq="out/d.dat", target="out/e.dat")
For convenience, a _make.R file is provided, containing these make() calls.
After
files <- dir(recursive=TRUE) files <- files[files != "_make.R"] files <- c(grep("/", files, value=TRUE), grep("/", files, value=TRUE, invert=TRUE)) cat(files, sep="\n")
dag_targets
knitr::opts_knit$set(root.dir="examples/dag_targets")

DAG example based on the example from the targets user
manual.
The second_target depends on first_target and outer_function, which in
turn depends on inner_function and global_object.
Before
cat(dir()[dir() != "_make.R"], sep="\n")
Run
make("first_target.R", NULL, "output/first_target.dat") make("global_object.R", NULL, "output/global_object.dat") make("second_target.R", prereq=c("output/first_target.dat", "output/global_object.dat", "inner_function.R", "outer_function.R"), target="output/second_target.dat")
For convenience, a _make.R file is provided, containing these make() calls.
After
files <- dir(recursive=TRUE) files <- files[files != "_make.R"] files <- c(grep("/", files, value=TRUE), grep("/", files, value=TRUE, invert=TRUE)) cat(files, sep="\n")
Discussion
Use cases
The make() function is a tool that can be applied to many types of workflows,
consisting of one or many R scripts. It is especially useful when the complete
workflow takes many minutes or hours to run. Changing one part of the analysis
will then update the related plots and tables, without rerunning every part of
the analysis.
Your project
Most analyses resemble the sequential example above, dividing the workflow
into steps that run one after another. As an introductory example, the
sequential workflow consists of only three steps: model, plots, and tables.
In practice, it is usually practical to divide a workflow into more steps than that, the first step being data preparation, such as importing, filtering, aggregating, and converting the data to the format that the model expects.
If the model is non-trivial, it can be practical to have an output.R step righter after model.R, extracting the results from the model-specific format to a more general format that is easy to browse, tabulate, and plot. This way, the model.R script can be very short, making it easy to see and understand the modelling approach and configuration. Separating the fundamental modelling step from the manual labor of data preparation and plotting can make an analysis more open and reproducible - for others to browse and reuse.
The paradigm of using small dedicated scripts with clear input and output files (read and write function calls near the beginning and end of each script) is usually a better workflow design than managing a large monolithic script where the user navigates between sections to run selected blocks of code.
Comparison with other packages
The four_minutes and dag_targets examples above provide an interesting
comparison between the makeit package and the targets package, for example.
makeit
The makeit package is script-based, where each step passes the results to the
next step as output files. The user organizes their workflow by writing scripts
that produce files.
The makeit package has no package dependencies, takes a very short time to
learn, and can be used to run any existing workflows, as long as they are based
on scripts with input and output files. The scripts may include functions, but
that is not a requirement.
The package consists of a single function that does one thing: run an R script if underlying files have changed, otherwise do nothing.
TAF
The TAF package contains a similar make() function and is an ancestor of the
makeit package. The overall aim of TAF is to support and strengthen
reproducibility in science, as well as reviewability.
The TAF package provides a structured modular design that divides a workflow
into four main stages: data.R, model.R, output.R, and report.R. The
initial data are declared in a DATA.bib file and an optional SOFTWARE.bib
file can be used to declare specific versions of R packages and other software.
The package consists of many useful tools to support reproducible workflows for scientific analyses.
targets
The targets package is function-based, where each step passes the results to
the next step as objects in memory. The user organizes their workflow by writing
functions that produce objects. It is the successor of the older drake
package.
The targets package relies on many underlying packages, takes some time to
learn, and some work may be required to realign existing workflows into
functions. The functions may produce files, but that is not a requirement.
The package consists of many useful tools to support workflow design and management.
Comparison
Package | Paradigm | State | Package dependencies | Time to learn | Run existing workflow | Features
--------- | --------- | ------ | -------------------- | ------------- | ---------------------- | --------
makeit | Scripts | Files | None | Very short | Must be file-based | One
TAF | Scripts | Files | None | Some | Must be file-based | Many
targets | Functions | Memory | Many | Some | Must be function-based | Many
The CRAN task view for reproducible research provides an annotated list of packages related to pipeline toolkits and project workflows.
References
Magnusson, A. makeit: Run R Scripts if Needed.\ https://cran.r-project.org/package=makeit
Magnusson, A. and C. Millar. TAF: Transparent Assessment Framework for Reproducible Research.\ https://cran.r-project.org/package=TAF
Landau, W.M. 2021. The targets R package: a dynamic Make-like function-oriented pipeline toolkit for reproducibility and high-performance computing. Journal of Open Source Software, 6(57), 2959.\ https://doi.org/10.21105/joss.02959, https://cran.r-project.org/package=targets
Stallman, R.M. et al. An introduction to makefiles. Chapter 2 in the GNU Make manual.\ https://www.gnu.org/software/make/manual/
Try the makeit package in your browser
Any scripts or data that you put into this service are public.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
