Introduction to metalite.sl
In metalite.sl: Subject-Level Analysis Using 'metalite'
knitr::opts_chunk$set(
comment = "#>",
collapse = TRUE,
out.width = "100%",
dpi = 150,
eval = TRUE
)
library(metalite)
library(metalite.sl)
Overview
metalite.sl R package designed for the analysis & reporting of subject-level analysis in clinical trials.
It operates on ADaM datasets and adheres to the metalite structure.
The package encompasses the following components:
Baseline Characteristics.
This R package offers a comprehensive software development lifecycle (SDLC)
solution, encompassing activities such as definition, development,
validation, and finalization of the analysis.
Highlighted features
- Avoid duplicated input by using metadata structure.
- For example, define analysis population once to use
in all subject level analysis.
- Consistent input and output in standard functions.
- Provide workflow to add interactive features to subject-level analysis table.
Workflow
The overall workflow includes the following steps:
- Define metadata information using metalite R package.
- Prepare outdata using
prepare_*() functions.
- Extend outdata using
extend_*() functions (optional).
- Format outdata using
format_*() functions.
- Create TLFs using
tlf_*() functions.
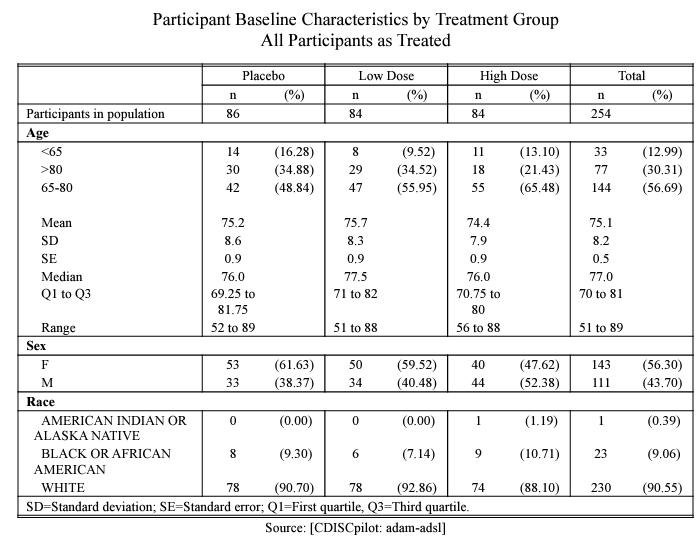
For instance, we can illustrate the creation of a straightforward
Baseline characteristic table as shown below.
meta_sl_example() |>
prepare_base_char(
population = "apat",
analysis = "base_char",
parameter = "age;gender"
) |>
format_base_char() |>
rtf_base_char(
source = "Source: [CDISCpilot: adam-adsl]",
path_outdata = tempfile(fileext = ".Rdata"),
path_outtable = tempfile(fileext = ".rtf")
)
knitr::include_graphics("pdf/base0char.pdf")
An example for interactive baseline characteristic table:
react_base_char(
metadata_sl = meta_sl_example(),
metadata_ae = metalite.ae::meta_ae_example(),
population = "apat",
observation = "wk12",
display_total = TRUE,
sl_parameter = "age;race",
ae_subgroup = c("age", "race"),
ae_specific = "rel",
width = 1200
)
Additional examples and tutorials can be found on the
package website,
offering further guidance and illustrations.
Input
To implement the workflow in metalite.sl, it is necessary to establish
a metadata structure using the metalite R package.
For detailed instructions, please consult the
metalite tutorial
and refer to the source code of the function
meta_sl_example().
Try the metalite.sl package in your browser
Any scripts or data that you put into this service are public.
metalite.sl documentation built on April 3, 2025, 8:52 p.m.
knitr::opts_chunk$set( comment = "#>", collapse = TRUE, out.width = "100%", dpi = 150, eval = TRUE )
library(metalite) library(metalite.sl)
Overview
metalite.sl R package designed for the analysis & reporting of subject-level analysis in clinical trials. It operates on ADaM datasets and adheres to the metalite structure. The package encompasses the following components:
Baseline Characteristics.

This R package offers a comprehensive software development lifecycle (SDLC) solution, encompassing activities such as definition, development, validation, and finalization of the analysis.
Highlighted features
- Avoid duplicated input by using metadata structure.
- For example, define analysis population once to use in all subject level analysis.
- Consistent input and output in standard functions.
- Provide workflow to add interactive features to subject-level analysis table.
Workflow
The overall workflow includes the following steps:
- Define metadata information using metalite R package.
- Prepare outdata using
prepare_*()functions. - Extend outdata using
extend_*()functions (optional). - Format outdata using
format_*()functions. - Create TLFs using
tlf_*()functions.
For instance, we can illustrate the creation of a straightforward Baseline characteristic table as shown below.
meta_sl_example() |> prepare_base_char( population = "apat", analysis = "base_char", parameter = "age;gender" ) |> format_base_char() |> rtf_base_char( source = "Source: [CDISCpilot: adam-adsl]", path_outdata = tempfile(fileext = ".Rdata"), path_outtable = tempfile(fileext = ".rtf") )
knitr::include_graphics("pdf/base0char.pdf")
An example for interactive baseline characteristic table:
react_base_char( metadata_sl = meta_sl_example(), metadata_ae = metalite.ae::meta_ae_example(), population = "apat", observation = "wk12", display_total = TRUE, sl_parameter = "age;race", ae_subgroup = c("age", "race"), ae_specific = "rel", width = 1200 )
Additional examples and tutorials can be found on the package website, offering further guidance and illustrations.
Input
To implement the workflow in metalite.sl, it is necessary to establish
a metadata structure using the metalite R package.
For detailed instructions, please consult the
metalite tutorial
and refer to the source code of the function
meta_sl_example().
Try the metalite.sl package in your browser
Any scripts or data that you put into this service are public.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
