README.md
In DHLab-TSENG/lab: a package for EHR laboratory data
Getting started with lab package
For full references, please refer to:
https://dhlab-tseng.github.io/lab/index.html

I. Introduction
The proposed open-source lab package is a software tool that help users
to explore and process laboratory data in electronic health records
(EHRs). With the lab package, researchers can easily map local
laboratory codes to the universal standard, mark abnormal results,
summarize data using descriptive statistics, impute missing values, and
generate analysis ready data.
Feature
- Data Mapping Standardize and manipulate data with Logical
Observation Identifiers Names and Codes (LOINC), a common
terminology for laboratory and clinical observations.
- Time Series Analysis Separate lab test results into multiple
consecutive non-overlapped time windows
- Value Imputation Impute value to replace missing data
- Wide Format Generation Transform longitudinal data into wide
format to generate analysis ready data
Development version
# install.packages("remotes")
remotes::install_github("DHLab-TSENG/lab")
Overview
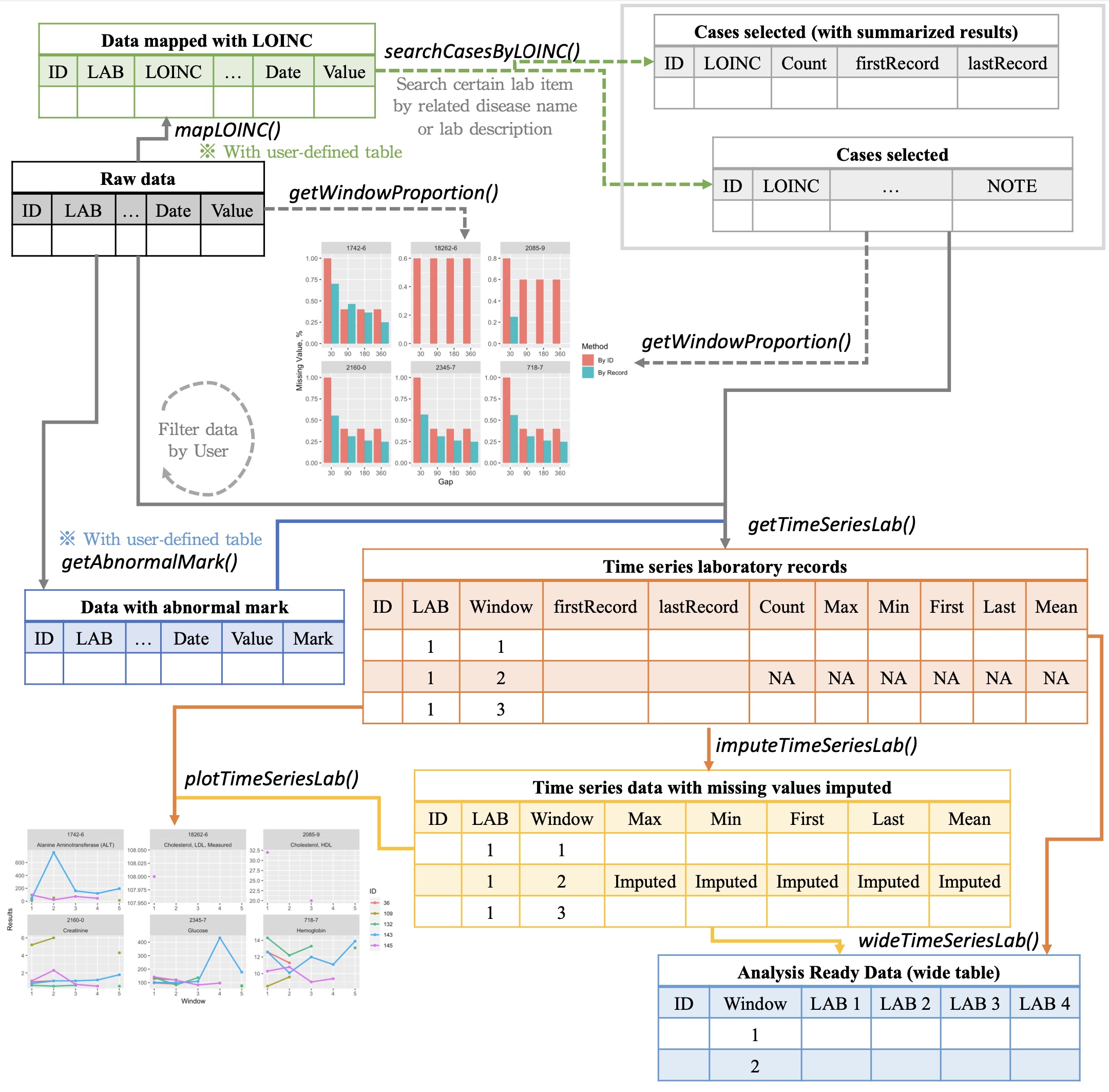
Usage
# install.packages("remotes")
remotes::install_github("DHLab-TSENG/lab")
library(lab)
Dataset
The sample data includes 1,744 lab records containing 7 different lab
items tested by 5 patients from MIMIC-III database.
head(labSample)
#> SUBJECT_ID ITEMID CHARTTIME VALUENUM VALUEUOM FLAG
#> 1: 36 50811 2131-05-18 12.7 g/dL abnormal
#> 2: 36 50912 2131-05-18 1.2 mg/dL
#> 3: 36 51222 2131-05-18 11.9 g/dL abnormal
#> 4: 36 50912 2131-05-19 1.3 mg/dL abnormal
#> 5: 36 50931 2131-05-19 160.0 mg/dL abnormal
#> 6: 36 51222 2131-05-19 9.6 g/dL abnormal
I. Data Mapping
If LOINC is not the default terminology, users are recommended to map
local lab item with LOINC by providing mapping table.
First, user shall prepare a mapping table with local codes and LOINC
codes.
head(mapSample)
#> ITEMID LABEL FLUID CATEGORY LOINC
#> 1: 50811 Hemoglobin Blood Blood Gas 718-7
#> 2: 50861 Alanine Aminotransferase (ALT) Blood Chemistry 1742-6
#> 3: 50904 Cholesterol, HDL Blood Chemistry 2085-9
#> 4: 50906 Cholesterol, LDL, Measured Blood Chemistry 18262-6
#> 5: 50912 Creatinine Blood Chemistry 2160-0
#> 6: 50931 Glucose Blood Chemistry 2345-7
loincSample <- mapLOINC(labData = labSample, labItemColName = ITEMID, mappingTable = mapSample)
head(loincSample)
#> ITEMID SUBJECT_ID CHARTTIME VALUENUM VALUEUOM FLAG LABEL FLUID
#> 1: 50811 36 2131-05-18 12.7 g/dL abnormal Hemoglobin Blood
#> 2: 50811 36 2131-05-04 12.3 g/dL abnormal Hemoglobin Blood
#> 3: 50811 36 2131-05-15 10.0 g/dL abnormal Hemoglobin Blood
#> 4: 50811 36 2131-05-17 11.7 g/dL abnormal Hemoglobin Blood
#> 5: 50811 109 2142-02-25 6.9 g/dL abnormal Hemoglobin Blood
#> 6: 50811 109 2141-09-20 7.2 g/dL abnormal Hemoglobin Blood
#> CATEGORY LOINC
#> 1: Blood Gas 718-7
#> 2: Blood Gas 718-7
#> 3: Blood Gas 718-7
#> 4: Blood Gas 718-7
#> 5: Blood Gas 718-7
#> 6: Blood Gas 718-7
Once a user map lab test codes with LOINC, ranges can be used to mark
abnormal results, and related names can be used to search related lab
test codes by other common names of a lab test.
loincMarkedSample <- getAbnormalMark(labData = loincSample,
idColName = SUBJECT_ID,
labItemColName = LOINC,
valueColName = VALUENUM,
genderColName = GENDER,
genderTable = patientSample,
referenceTable = refLOINC)
head(loincMarkedSample)
#> ITEMID ID CHARTTIME Value VALUEUOM FLAG LABEL
#> 1: 50861 36 2131-04-30 8 IU/L Alanine Aminotransferase (ALT)
#> 2: 50861 36 2131-05-17 12 IU/L Alanine Aminotransferase (ALT)
#> 3: 50861 36 2134-05-14 12 IU/L Alanine Aminotransferase (ALT)
#> 4: 50861 109 2138-07-03 14 IU/L Alanine Aminotransferase (ALT)
#> 5: 50861 109 2142-03-21 46 IU/L abnormal Alanine Aminotransferase (ALT)
#> 6: 50861 109 2142-01-09 10 IU/L Alanine Aminotransferase (ALT)
#> FLUID CATEGORY LOINC ABMark
#> 1: Blood Chemistry 1742-6 <NA>
#> 2: Blood Chemistry 1742-6 <NA>
#> 3: Blood Chemistry 1742-6 <NA>
#> 4: Blood Chemistry 1742-6 <NA>
#> 5: Blood Chemistry 1742-6 H
#> 6: Blood Chemistry 1742-6 <NA>
caseCreatinine <- searchCasesByLOINC(labData = loincSample,
idColName = SUBJECT_ID,
loincColName = LOINC,
dateColName = CHARTTIME,
condition = "Creatinine",
isSummary = TRUE)
head(caseCreatinine)
#> ID LOINC Count firstRecord lastRecode
#> 1: 36 2160-0 37 2131-04-30 2134-05-20
#> 2: 109 2160-0 238 2137-11-04 2142-08-30
#> 3: 132 2160-0 32 2115-05-06 2116-04-08
#> 4: 143 2160-0 60 2154-12-25 2155-10-22
#> 5: 145 2160-0 162 2144-03-29 2145-02-22
II. Time Series Analysis
lab package allows users to separate lab test results into multiple
consecutive non-overlapped time windows. The index date of time windows
can be the first or last event occurred for individuals, or a specific
date for all patients. To help users find suitable window size (e.g., 30
days or 180 days, to name but a few), a plot function is provided to
visualize how frequent the patients did each lab test.
windowProportion <- plotWindowProportion(labData = loincSample,
idColName = SUBJECT_ID,
labItemColName = LOINC,
dateColName = CHARTTIME,
indexDate = first,
gapDate = c(30, 90, 180, 360),
studyPeriodStartDays=0,
studyPeriodEndDays=360)
print(windowProportion$graph)

head(windowProportion$missingData)
#> LAB Gap Method Proportion
#> 1: 1742-6 30 By Individual 0
#> 2: 1742-6 30 By Individual 0
#> 3: 1742-6 30 By Individual 0
#> 4: 1742-6 30 By Individual 0
#> 5: 1742-6 30 By Individual 0
#> 6: 2160-0 30 By Individual 0
After the index date and window size are decided, the descriptive
statistics information, including total test times within a window,
maximum test value, minimum test value, test values average, and the
record nearest to the index date, are shown.
timeSeriesData <- getTimeSeriesLab(labData = loincSample,
idColName = SUBJECT_ID,
labItemColName = LOINC + LABEL,
dateColName = CHARTTIME,
valueColName = VALUENUM,
indexDate = first,
gapDate = 30,
completeWindows = TRUE)
head(timeSeriesData)
#> ID LOINC LABEL Window Count Max Min Mean Nearest
#> 1: 36 1742-6 Alanine Aminotransferase (ALT) 1 2 12 8 10 8
#> 2: 36 1742-6 Alanine Aminotransferase (ALT) 2 NA NA NA NA NA
#> 3: 36 1742-6 Alanine Aminotransferase (ALT) 3 NA NA NA NA NA
#> 4: 36 1742-6 Alanine Aminotransferase (ALT) 4 NA NA NA NA NA
#> 5: 36 1742-6 Alanine Aminotransferase (ALT) 5 NA NA NA NA NA
#> 6: 36 1742-6 Alanine Aminotransferase (ALT) 6 NA NA NA NA NA
#> firstRecord lastRecode
#> 1: 2131-04-30 2131-05-17
#> 2: <NA> <NA>
#> 3: <NA> <NA>
#> 4: <NA> <NA>
#> 5: <NA> <NA>
#> 6: <NA> <NA>
Also, a line chart plotting function is available to do long-term
follow-up. Visualization is helpful for detecting data trends.
Additionally, “L” and “H” will be used as legendary icon if abnormal
values are marked.
timeSeriesPlot <- plotTimeSeriesLab(labData = timeSeriesData,
idColName = ID,
labItemColName = LOINC + LABEL,
timeMarkColName = Window,
valueColName = Nearest,
timeStart = 1,
timeEnd = 5,
abnormalMarkColName = NULL)
plot(timeSeriesPlot)

III. Imputation
Imputation function can be executed to replace missing data.
fullTimeSeriesData <- imputeTimeSeriesLab(labData = timeSeriesData,
idColName = ID,
labItemColName = LOINC + LABEL,
windowColName = Window,
valueColName = Mean & Nearest,
impMethod = NOCB,
imputeOverallMean = FALSE)
fullTimeSeriesData[timeSeriesData$ID==36&
timeSeriesData$LOINC=="2160-0"]
#> ID LOINC LABEL Window Mean Nearest imputed
#> 1: 36 2160-0 Creatinine 1 1.2347826 1.0 FALSE
#> 2: 36 2160-0 Creatinine 2 1.1000000 1.1 FALSE
#> 3: 36 2160-0 Creatinine 3 1.1000000 1.1 TRUE
#> 4: 36 2160-0 Creatinine 4 1.1000000 1.1 TRUE
#> 5: 36 2160-0 Creatinine 5 1.1000000 1.1 TRUE
#> 6: 36 2160-0 Creatinine 6 1.1000000 1.1 TRUE
#> 7: 36 2160-0 Creatinine 7 1.1000000 1.1 TRUE
#> 8: 36 2160-0 Creatinine 8 1.1000000 1.1 TRUE
#> 9: 36 2160-0 Creatinine 9 1.2000000 1.2 FALSE
#> 10: 36 2160-0 Creatinine 10 1.1000000 1.1 FALSE
#> 11: 36 2160-0 Creatinine 11 1.1000000 1.1 TRUE
#> 12: 36 2160-0 Creatinine 12 1.1000000 1.1 TRUE
#> 13: 36 2160-0 Creatinine 13 1.1000000 1.1 TRUE
#> 14: 36 2160-0 Creatinine 14 1.1000000 1.1 TRUE
#> 15: 36 2160-0 Creatinine 15 1.1000000 1.1 TRUE
#> 16: 36 2160-0 Creatinine 16 1.1000000 1.1 TRUE
#> 17: 36 2160-0 Creatinine 17 1.1000000 1.1 TRUE
#> 18: 36 2160-0 Creatinine 18 1.1000000 1.1 TRUE
#> 19: 36 2160-0 Creatinine 19 1.1000000 1.1 TRUE
#> 20: 36 2160-0 Creatinine 20 1.1000000 1.1 TRUE
#> 21: 36 2160-0 Creatinine 21 1.1000000 1.1 TRUE
#> 22: 36 2160-0 Creatinine 22 1.1000000 1.1 TRUE
#> 23: 36 2160-0 Creatinine 23 1.1000000 1.1 TRUE
#> 24: 36 2160-0 Creatinine 24 1.1000000 1.1 TRUE
#> 25: 36 2160-0 Creatinine 25 1.1000000 1.1 TRUE
#> 26: 36 2160-0 Creatinine 26 1.1000000 1.1 TRUE
#> 27: 36 2160-0 Creatinine 27 1.1000000 1.1 TRUE
#> 28: 36 2160-0 Creatinine 28 1.1000000 1.1 TRUE
#> 29: 36 2160-0 Creatinine 29 1.1000000 1.1 TRUE
#> 30: 36 2160-0 Creatinine 30 1.1000000 1.1 TRUE
#> 31: 36 2160-0 Creatinine 31 1.1000000 1.1 TRUE
#> 32: 36 2160-0 Creatinine 32 1.1000000 1.1 TRUE
#> 33: 36 2160-0 Creatinine 33 1.1000000 1.1 TRUE
#> 34: 36 2160-0 Creatinine 34 1.1000000 1.1 TRUE
#> 35: 36 2160-0 Creatinine 35 1.1000000 1.1 TRUE
#> 36: 36 2160-0 Creatinine 36 1.1000000 1.1 TRUE
#> 37: 36 2160-0 Creatinine 37 0.9666667 1.0 FALSE
#> 38: 36 2160-0 Creatinine 38 0.8500000 0.9 FALSE
#> ID LOINC LABEL Window Mean Nearest imputed
IV. Wide Format Generation
Then, a function be used to transform longitudinal data into wide format
to generate analysis ready data.
wideTimeSeriesData <- wideTimeSeriesLab(labData = fullTimeSeriesData,
idColName = ID,
labItemColName = LOINC + LABEL,
windowColName = Window,
valueColName = Nearest)
head(wideTimeSeriesData)
#> ID Window 1742-6_Alanine Aminotransferase (ALT)
#> 1: 36 1 8
#> 2: 36 2 8
#> 3: 36 3 8
#> 4: 36 4 8
#> 5: 36 5 8
#> 6: 36 6 8
#> 18262-6_Cholesterol, LDL, Measured 2085-9_Cholesterol, HDL 2160-0_Creatinine
#> 1: NA NA 1.0
#> 2: NA NA 1.1
#> 3: NA NA 1.1
#> 4: NA NA 1.1
#> 5: NA NA 1.1
#> 6: NA NA 1.1
#> 2345-7_Glucose 718-7_Hemoglobin
#> 1: 98 12.6
#> 2: 90 11.3
#> 3: 90 11.3
#> 4: 90 11.3
#> 5: 90 11.3
#> 6: 90 11.3
V. Machine Learning Application
Wide format date is commonly utilized in machine learning methods. In
this package, we provide an k-nearest neighbors (kNN) imputation
function enabling users to impute missing values by machine learning
technique with wide format data.
wideTimeSeriesData <- wideTimeSeriesLab(labData = timeSeriesData,
idColName = ID,
labItemColName = LOINC + LABEL,
windowColName = Window,
valueColName = Nearest)
knnImputedData <- imputeKNN(labData = wideTimeSeriesData,
idColName = ID + Window,
k = 1)
head(knnImputedData)
#> ID Window 1742-6_Alanine Aminotransferase (ALT)
#> 1: 36 1 8
#> 2: 36 2 8
#> 3: 36 3 8
#> 4: 36 4 8
#> 5: 36 5 8
#> 6: 36 6 8
#> 18262-6_Cholesterol, LDL, Measured 2085-9_Cholesterol, HDL 2160-0_Creatinine
#> 1: 108 32 1.0
#> 2: 108 32 1.1
#> 3: 108 32 1.1
#> 4: 108 32 1.1
#> 5: 108 32 1.1
#> 6: 108 32 1.1
#> 2345-7_Glucose 718-7_Hemoglobin
#> 1: 98 12.6
#> 2: 90 11.3
#> 3: 90 11.3
#> 4: 90 11.3
#> 5: 90 11.3
#> 6: 90 11.3
Citation
Tseng YJ, Chen CJ, Chang CW. 2023. lab: an R package for generating analysis-ready data from laboratory records. PeerJ Computer Science 9:e1528 https://doi.org/10.7717/peerj-cs.1528
DHLab-TSENG/lab documentation built on Sept. 1, 2023, 9:03 p.m.
Getting started with lab package
For full references, please refer to: https://dhlab-tseng.github.io/lab/index.html

I. Introduction
The proposed open-source lab package is a software tool that help users to explore and process laboratory data in electronic health records (EHRs). With the lab package, researchers can easily map local laboratory codes to the universal standard, mark abnormal results, summarize data using descriptive statistics, impute missing values, and generate analysis ready data.
Feature
- Data Mapping Standardize and manipulate data with Logical Observation Identifiers Names and Codes (LOINC), a common terminology for laboratory and clinical observations.
- Time Series Analysis Separate lab test results into multiple consecutive non-overlapped time windows
- Value Imputation Impute value to replace missing data
- Wide Format Generation Transform longitudinal data into wide format to generate analysis ready data
Development version
# install.packages("remotes")
remotes::install_github("DHLab-TSENG/lab")
Overview

Usage
# install.packages("remotes")
remotes::install_github("DHLab-TSENG/lab")
library(lab)
Dataset
The sample data includes 1,744 lab records containing 7 different lab items tested by 5 patients from MIMIC-III database.
head(labSample)
#> SUBJECT_ID ITEMID CHARTTIME VALUENUM VALUEUOM FLAG
#> 1: 36 50811 2131-05-18 12.7 g/dL abnormal
#> 2: 36 50912 2131-05-18 1.2 mg/dL
#> 3: 36 51222 2131-05-18 11.9 g/dL abnormal
#> 4: 36 50912 2131-05-19 1.3 mg/dL abnormal
#> 5: 36 50931 2131-05-19 160.0 mg/dL abnormal
#> 6: 36 51222 2131-05-19 9.6 g/dL abnormal
I. Data Mapping
If LOINC is not the default terminology, users are recommended to map local lab item with LOINC by providing mapping table.
First, user shall prepare a mapping table with local codes and LOINC codes.
head(mapSample)
#> ITEMID LABEL FLUID CATEGORY LOINC
#> 1: 50811 Hemoglobin Blood Blood Gas 718-7
#> 2: 50861 Alanine Aminotransferase (ALT) Blood Chemistry 1742-6
#> 3: 50904 Cholesterol, HDL Blood Chemistry 2085-9
#> 4: 50906 Cholesterol, LDL, Measured Blood Chemistry 18262-6
#> 5: 50912 Creatinine Blood Chemistry 2160-0
#> 6: 50931 Glucose Blood Chemistry 2345-7
loincSample <- mapLOINC(labData = labSample, labItemColName = ITEMID, mappingTable = mapSample)
head(loincSample)
#> ITEMID SUBJECT_ID CHARTTIME VALUENUM VALUEUOM FLAG LABEL FLUID
#> 1: 50811 36 2131-05-18 12.7 g/dL abnormal Hemoglobin Blood
#> 2: 50811 36 2131-05-04 12.3 g/dL abnormal Hemoglobin Blood
#> 3: 50811 36 2131-05-15 10.0 g/dL abnormal Hemoglobin Blood
#> 4: 50811 36 2131-05-17 11.7 g/dL abnormal Hemoglobin Blood
#> 5: 50811 109 2142-02-25 6.9 g/dL abnormal Hemoglobin Blood
#> 6: 50811 109 2141-09-20 7.2 g/dL abnormal Hemoglobin Blood
#> CATEGORY LOINC
#> 1: Blood Gas 718-7
#> 2: Blood Gas 718-7
#> 3: Blood Gas 718-7
#> 4: Blood Gas 718-7
#> 5: Blood Gas 718-7
#> 6: Blood Gas 718-7
Once a user map lab test codes with LOINC, ranges can be used to mark abnormal results, and related names can be used to search related lab test codes by other common names of a lab test.
loincMarkedSample <- getAbnormalMark(labData = loincSample,
idColName = SUBJECT_ID,
labItemColName = LOINC,
valueColName = VALUENUM,
genderColName = GENDER,
genderTable = patientSample,
referenceTable = refLOINC)
head(loincMarkedSample)
#> ITEMID ID CHARTTIME Value VALUEUOM FLAG LABEL
#> 1: 50861 36 2131-04-30 8 IU/L Alanine Aminotransferase (ALT)
#> 2: 50861 36 2131-05-17 12 IU/L Alanine Aminotransferase (ALT)
#> 3: 50861 36 2134-05-14 12 IU/L Alanine Aminotransferase (ALT)
#> 4: 50861 109 2138-07-03 14 IU/L Alanine Aminotransferase (ALT)
#> 5: 50861 109 2142-03-21 46 IU/L abnormal Alanine Aminotransferase (ALT)
#> 6: 50861 109 2142-01-09 10 IU/L Alanine Aminotransferase (ALT)
#> FLUID CATEGORY LOINC ABMark
#> 1: Blood Chemistry 1742-6 <NA>
#> 2: Blood Chemistry 1742-6 <NA>
#> 3: Blood Chemistry 1742-6 <NA>
#> 4: Blood Chemistry 1742-6 <NA>
#> 5: Blood Chemistry 1742-6 H
#> 6: Blood Chemistry 1742-6 <NA>
caseCreatinine <- searchCasesByLOINC(labData = loincSample,
idColName = SUBJECT_ID,
loincColName = LOINC,
dateColName = CHARTTIME,
condition = "Creatinine",
isSummary = TRUE)
head(caseCreatinine)
#> ID LOINC Count firstRecord lastRecode
#> 1: 36 2160-0 37 2131-04-30 2134-05-20
#> 2: 109 2160-0 238 2137-11-04 2142-08-30
#> 3: 132 2160-0 32 2115-05-06 2116-04-08
#> 4: 143 2160-0 60 2154-12-25 2155-10-22
#> 5: 145 2160-0 162 2144-03-29 2145-02-22
II. Time Series Analysis
lab package allows users to separate lab test results into multiple consecutive non-overlapped time windows. The index date of time windows can be the first or last event occurred for individuals, or a specific date for all patients. To help users find suitable window size (e.g., 30 days or 180 days, to name but a few), a plot function is provided to visualize how frequent the patients did each lab test.
windowProportion <- plotWindowProportion(labData = loincSample,
idColName = SUBJECT_ID,
labItemColName = LOINC,
dateColName = CHARTTIME,
indexDate = first,
gapDate = c(30, 90, 180, 360),
studyPeriodStartDays=0,
studyPeriodEndDays=360)
print(windowProportion$graph)

head(windowProportion$missingData)
#> LAB Gap Method Proportion
#> 1: 1742-6 30 By Individual 0
#> 2: 1742-6 30 By Individual 0
#> 3: 1742-6 30 By Individual 0
#> 4: 1742-6 30 By Individual 0
#> 5: 1742-6 30 By Individual 0
#> 6: 2160-0 30 By Individual 0
After the index date and window size are decided, the descriptive statistics information, including total test times within a window, maximum test value, minimum test value, test values average, and the record nearest to the index date, are shown.
timeSeriesData <- getTimeSeriesLab(labData = loincSample,
idColName = SUBJECT_ID,
labItemColName = LOINC + LABEL,
dateColName = CHARTTIME,
valueColName = VALUENUM,
indexDate = first,
gapDate = 30,
completeWindows = TRUE)
head(timeSeriesData)
#> ID LOINC LABEL Window Count Max Min Mean Nearest
#> 1: 36 1742-6 Alanine Aminotransferase (ALT) 1 2 12 8 10 8
#> 2: 36 1742-6 Alanine Aminotransferase (ALT) 2 NA NA NA NA NA
#> 3: 36 1742-6 Alanine Aminotransferase (ALT) 3 NA NA NA NA NA
#> 4: 36 1742-6 Alanine Aminotransferase (ALT) 4 NA NA NA NA NA
#> 5: 36 1742-6 Alanine Aminotransferase (ALT) 5 NA NA NA NA NA
#> 6: 36 1742-6 Alanine Aminotransferase (ALT) 6 NA NA NA NA NA
#> firstRecord lastRecode
#> 1: 2131-04-30 2131-05-17
#> 2: <NA> <NA>
#> 3: <NA> <NA>
#> 4: <NA> <NA>
#> 5: <NA> <NA>
#> 6: <NA> <NA>
Also, a line chart plotting function is available to do long-term follow-up. Visualization is helpful for detecting data trends. Additionally, “L” and “H” will be used as legendary icon if abnormal values are marked.
timeSeriesPlot <- plotTimeSeriesLab(labData = timeSeriesData,
idColName = ID,
labItemColName = LOINC + LABEL,
timeMarkColName = Window,
valueColName = Nearest,
timeStart = 1,
timeEnd = 5,
abnormalMarkColName = NULL)
plot(timeSeriesPlot)

III. Imputation
Imputation function can be executed to replace missing data.
fullTimeSeriesData <- imputeTimeSeriesLab(labData = timeSeriesData,
idColName = ID,
labItemColName = LOINC + LABEL,
windowColName = Window,
valueColName = Mean & Nearest,
impMethod = NOCB,
imputeOverallMean = FALSE)
fullTimeSeriesData[timeSeriesData$ID==36&
timeSeriesData$LOINC=="2160-0"]
#> ID LOINC LABEL Window Mean Nearest imputed
#> 1: 36 2160-0 Creatinine 1 1.2347826 1.0 FALSE
#> 2: 36 2160-0 Creatinine 2 1.1000000 1.1 FALSE
#> 3: 36 2160-0 Creatinine 3 1.1000000 1.1 TRUE
#> 4: 36 2160-0 Creatinine 4 1.1000000 1.1 TRUE
#> 5: 36 2160-0 Creatinine 5 1.1000000 1.1 TRUE
#> 6: 36 2160-0 Creatinine 6 1.1000000 1.1 TRUE
#> 7: 36 2160-0 Creatinine 7 1.1000000 1.1 TRUE
#> 8: 36 2160-0 Creatinine 8 1.1000000 1.1 TRUE
#> 9: 36 2160-0 Creatinine 9 1.2000000 1.2 FALSE
#> 10: 36 2160-0 Creatinine 10 1.1000000 1.1 FALSE
#> 11: 36 2160-0 Creatinine 11 1.1000000 1.1 TRUE
#> 12: 36 2160-0 Creatinine 12 1.1000000 1.1 TRUE
#> 13: 36 2160-0 Creatinine 13 1.1000000 1.1 TRUE
#> 14: 36 2160-0 Creatinine 14 1.1000000 1.1 TRUE
#> 15: 36 2160-0 Creatinine 15 1.1000000 1.1 TRUE
#> 16: 36 2160-0 Creatinine 16 1.1000000 1.1 TRUE
#> 17: 36 2160-0 Creatinine 17 1.1000000 1.1 TRUE
#> 18: 36 2160-0 Creatinine 18 1.1000000 1.1 TRUE
#> 19: 36 2160-0 Creatinine 19 1.1000000 1.1 TRUE
#> 20: 36 2160-0 Creatinine 20 1.1000000 1.1 TRUE
#> 21: 36 2160-0 Creatinine 21 1.1000000 1.1 TRUE
#> 22: 36 2160-0 Creatinine 22 1.1000000 1.1 TRUE
#> 23: 36 2160-0 Creatinine 23 1.1000000 1.1 TRUE
#> 24: 36 2160-0 Creatinine 24 1.1000000 1.1 TRUE
#> 25: 36 2160-0 Creatinine 25 1.1000000 1.1 TRUE
#> 26: 36 2160-0 Creatinine 26 1.1000000 1.1 TRUE
#> 27: 36 2160-0 Creatinine 27 1.1000000 1.1 TRUE
#> 28: 36 2160-0 Creatinine 28 1.1000000 1.1 TRUE
#> 29: 36 2160-0 Creatinine 29 1.1000000 1.1 TRUE
#> 30: 36 2160-0 Creatinine 30 1.1000000 1.1 TRUE
#> 31: 36 2160-0 Creatinine 31 1.1000000 1.1 TRUE
#> 32: 36 2160-0 Creatinine 32 1.1000000 1.1 TRUE
#> 33: 36 2160-0 Creatinine 33 1.1000000 1.1 TRUE
#> 34: 36 2160-0 Creatinine 34 1.1000000 1.1 TRUE
#> 35: 36 2160-0 Creatinine 35 1.1000000 1.1 TRUE
#> 36: 36 2160-0 Creatinine 36 1.1000000 1.1 TRUE
#> 37: 36 2160-0 Creatinine 37 0.9666667 1.0 FALSE
#> 38: 36 2160-0 Creatinine 38 0.8500000 0.9 FALSE
#> ID LOINC LABEL Window Mean Nearest imputed
IV. Wide Format Generation
Then, a function be used to transform longitudinal data into wide format to generate analysis ready data.
wideTimeSeriesData <- wideTimeSeriesLab(labData = fullTimeSeriesData,
idColName = ID,
labItemColName = LOINC + LABEL,
windowColName = Window,
valueColName = Nearest)
head(wideTimeSeriesData)
#> ID Window 1742-6_Alanine Aminotransferase (ALT)
#> 1: 36 1 8
#> 2: 36 2 8
#> 3: 36 3 8
#> 4: 36 4 8
#> 5: 36 5 8
#> 6: 36 6 8
#> 18262-6_Cholesterol, LDL, Measured 2085-9_Cholesterol, HDL 2160-0_Creatinine
#> 1: NA NA 1.0
#> 2: NA NA 1.1
#> 3: NA NA 1.1
#> 4: NA NA 1.1
#> 5: NA NA 1.1
#> 6: NA NA 1.1
#> 2345-7_Glucose 718-7_Hemoglobin
#> 1: 98 12.6
#> 2: 90 11.3
#> 3: 90 11.3
#> 4: 90 11.3
#> 5: 90 11.3
#> 6: 90 11.3
V. Machine Learning Application
Wide format date is commonly utilized in machine learning methods. In this package, we provide an k-nearest neighbors (kNN) imputation function enabling users to impute missing values by machine learning technique with wide format data.
wideTimeSeriesData <- wideTimeSeriesLab(labData = timeSeriesData,
idColName = ID,
labItemColName = LOINC + LABEL,
windowColName = Window,
valueColName = Nearest)
knnImputedData <- imputeKNN(labData = wideTimeSeriesData,
idColName = ID + Window,
k = 1)
head(knnImputedData)
#> ID Window 1742-6_Alanine Aminotransferase (ALT)
#> 1: 36 1 8
#> 2: 36 2 8
#> 3: 36 3 8
#> 4: 36 4 8
#> 5: 36 5 8
#> 6: 36 6 8
#> 18262-6_Cholesterol, LDL, Measured 2085-9_Cholesterol, HDL 2160-0_Creatinine
#> 1: 108 32 1.0
#> 2: 108 32 1.1
#> 3: 108 32 1.1
#> 4: 108 32 1.1
#> 5: 108 32 1.1
#> 6: 108 32 1.1
#> 2345-7_Glucose 718-7_Hemoglobin
#> 1: 98 12.6
#> 2: 90 11.3
#> 3: 90 11.3
#> 4: 90 11.3
#> 5: 90 11.3
#> 6: 90 11.3
Citation
Tseng YJ, Chen CJ, Chang CW. 2023. lab: an R package for generating analysis-ready data from laboratory records. PeerJ Computer Science 9:e1528 https://doi.org/10.7717/peerj-cs.1528
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
