README.md
In FrankBaut/RandomClustering: Random clusters generation on a lattice
RandomClustering
Generate a binary (0,1) lattice of size 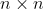 with parameter p, p indicates how many ones will be placed randomly with respect to the total lattice cells, this will gerate clusters of diferent sizes in the lattice.
with parameter p, p indicates how many ones will be placed randomly with respect to the total lattice cells, this will gerate clusters of diferent sizes in the lattice.
We follow the next steps:
-
Create a binary lattice of size 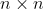 with a probability of filling
with a probability of filling 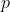 .
.
-
Label the clusters that are generated in the lattice.
-
Choose the biggets cluster formed in the lattice and randomly remove a cell.
-
Choose the smallest clusters and add randomly a cell with the same label of the selected cluster in one of his neighbors.
-
If there are several smallest clusters with the same size, we choose one of them randomly and we repeat the previous step.

Installation
# The development version from GitHub:
# install.packages("devtools")
devtools::install_github("FrankBaut/RandomClustering")
# install.packages("tidyverse")
# install.packages("magrittr")
Examples
# Basic workflow:
library("tidyverse")
library("magrittr")
library("RandomClustering")
Label_clusters <- clusters_matrix(n=100, p = 0.5) %>%
big_cl() %>% small_cl()
Maybe we would like observe the new clusters configuration after one iteration and re-label this configuration. Basically we must do:
Label_clusters <- clusters_matrix(n=100,p=0.5)
proceso(Label_clusters)
Or with pipe operator %>%:
Label_clusters <- clusters_matrix(n=10,p=0.5) %>% proceso()
Simulation
In the given case if you want to make a simulation and observe the biggets cluster in different p values, you can do this:
n<-100
iterations<-50
prob_vec<-seq(.01,.9,.01)
biggets_cluster<-matrix(,nrow = length(prob_vec),ncol = iterations)
for (i in 1:length(prob_vec)) {
data<-clusters_matrix(n,prob_vec[i])
for (j in 1:iterations) {
data<-proceso(data)
caso<-data %>% as.vector()%>% na.omit() %>% plyr::count()
biggets_cluster[i,j]<-max(caso$freq,na.rm = T)/sum(caso$freq,na.rm = T)
}
}
However this is really slow, therefore we must do a parallel process. Two libraries very useful are foreach
and doParallel.
# install.packages("foreach")
# install.packages("doParallel")
library(foreach)
library(doParallel)
library(RandomClustering)
iterations<-50
n<-100
prob_vec<-seq(.01,.9,.01)
#------------Progress bar------------------
l<-length(prob_vec)
pb <- txtProgressBar(0,l, style = 3)
#------------------------------------------
registerDoParallel(detectCores()-2)
biggets_cluster<-unlist(complete_processPar(n, prob_vec,iterations,l,pb))
stopImplicitCluster()
df<-data.frame(prob_vec,biggets_cluster)
plot(df,xlab="p",ylab="Biggest Cluster")

We can choose how many cores we want to use in registerDoParallel argument. we recommend using at most detectCores()-1 cores. Remenber stop the parallel process with stopImplicitCluster() when you finish your calculations.
If we want see how many clusters there are after several iterations we must do:
library(foreach)
library(doParallel)
library(RandomClustering)
iterations<-50
n<-100
prob_vec<-seq(.01,.9,.01)
#------------Progress bar------------------
l<-length(prob_vec)
pb <- txtProgressBar(0,l, style = 3)
#------------------------------------------
registerDoParallel(detectCores()-2)
biggets_cluster<-unlist(complete_clusterNumberPar(n, prob_vec,iterations,l,pb))
stopImplicitCluster()
df<-data.frame(prob_vec,biggets_cluster)
plot(df,xlab="p",ylab="Clusters number")

Please cite this work with:
citation("RandomClustering")
or in bibtex format
toBibtex(citation("RandomClustering"))
FrankBaut/RandomClustering documentation built on Feb. 19, 2020, 2:07 a.m.
RandomClustering
Generate a binary (0,1) lattice of size with parameter p, p indicates how many ones will be placed randomly with respect to the total lattice cells, this will gerate clusters of diferent sizes in the lattice.
We follow the next steps:
-
Create a binary lattice of size
with a probability of filling
.
-
Label the clusters that are generated in the lattice.
-
Choose the biggets cluster formed in the lattice and randomly remove a cell.
-
Choose the smallest clusters and add randomly a cell with the same label of the selected cluster in one of his neighbors.
-
If there are several smallest clusters with the same size, we choose one of them randomly and we repeat the previous step.

Installation
# The development version from GitHub:
# install.packages("devtools")
devtools::install_github("FrankBaut/RandomClustering")
# install.packages("tidyverse")
# install.packages("magrittr")
Examples
# Basic workflow:
library("tidyverse")
library("magrittr")
library("RandomClustering")
Label_clusters <- clusters_matrix(n=100, p = 0.5) %>%
big_cl() %>% small_cl()
Maybe we would like observe the new clusters configuration after one iteration and re-label this configuration. Basically we must do:
Label_clusters <- clusters_matrix(n=100,p=0.5)
proceso(Label_clusters)
Or with pipe operator %>%:
Label_clusters <- clusters_matrix(n=10,p=0.5) %>% proceso()
Simulation
In the given case if you want to make a simulation and observe the biggets cluster in different p values, you can do this:
n<-100
iterations<-50
prob_vec<-seq(.01,.9,.01)
biggets_cluster<-matrix(,nrow = length(prob_vec),ncol = iterations)
for (i in 1:length(prob_vec)) {
data<-clusters_matrix(n,prob_vec[i])
for (j in 1:iterations) {
data<-proceso(data)
caso<-data %>% as.vector()%>% na.omit() %>% plyr::count()
biggets_cluster[i,j]<-max(caso$freq,na.rm = T)/sum(caso$freq,na.rm = T)
}
}
However this is really slow, therefore we must do a parallel process. Two libraries very useful are foreach
and doParallel.
# install.packages("foreach")
# install.packages("doParallel")
library(foreach)
library(doParallel)
library(RandomClustering)
iterations<-50
n<-100
prob_vec<-seq(.01,.9,.01)
#------------Progress bar------------------
l<-length(prob_vec)
pb <- txtProgressBar(0,l, style = 3)
#------------------------------------------
registerDoParallel(detectCores()-2)
biggets_cluster<-unlist(complete_processPar(n, prob_vec,iterations,l,pb))
stopImplicitCluster()
df<-data.frame(prob_vec,biggets_cluster)
plot(df,xlab="p",ylab="Biggest Cluster")

We can choose how many cores we want to use in registerDoParallel argument. we recommend using at most detectCores()-1 cores. Remenber stop the parallel process with stopImplicitCluster() when you finish your calculations.
If we want see how many clusters there are after several iterations we must do:
library(foreach)
library(doParallel)
library(RandomClustering)
iterations<-50
n<-100
prob_vec<-seq(.01,.9,.01)
#------------Progress bar------------------
l<-length(prob_vec)
pb <- txtProgressBar(0,l, style = 3)
#------------------------------------------
registerDoParallel(detectCores()-2)
biggets_cluster<-unlist(complete_clusterNumberPar(n, prob_vec,iterations,l,pb))
stopImplicitCluster()
df<-data.frame(prob_vec,biggets_cluster)
plot(df,xlab="p",ylab="Clusters number")

Please cite this work with:
citation("RandomClustering")
or in bibtex format
toBibtex(citation("RandomClustering"))
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
