README.md
In Ghosn-Lab/BatchNorm: Data Matrix Normalization and Merging Strategies Minimize Batch-specific Systemic Variation in scRNA-Seq Data
Ghosn Laboratory 
Data Matrix Normalization and Merging Strategies Minimize Batch-specific Systemic Variation in scRNA-Seq Data
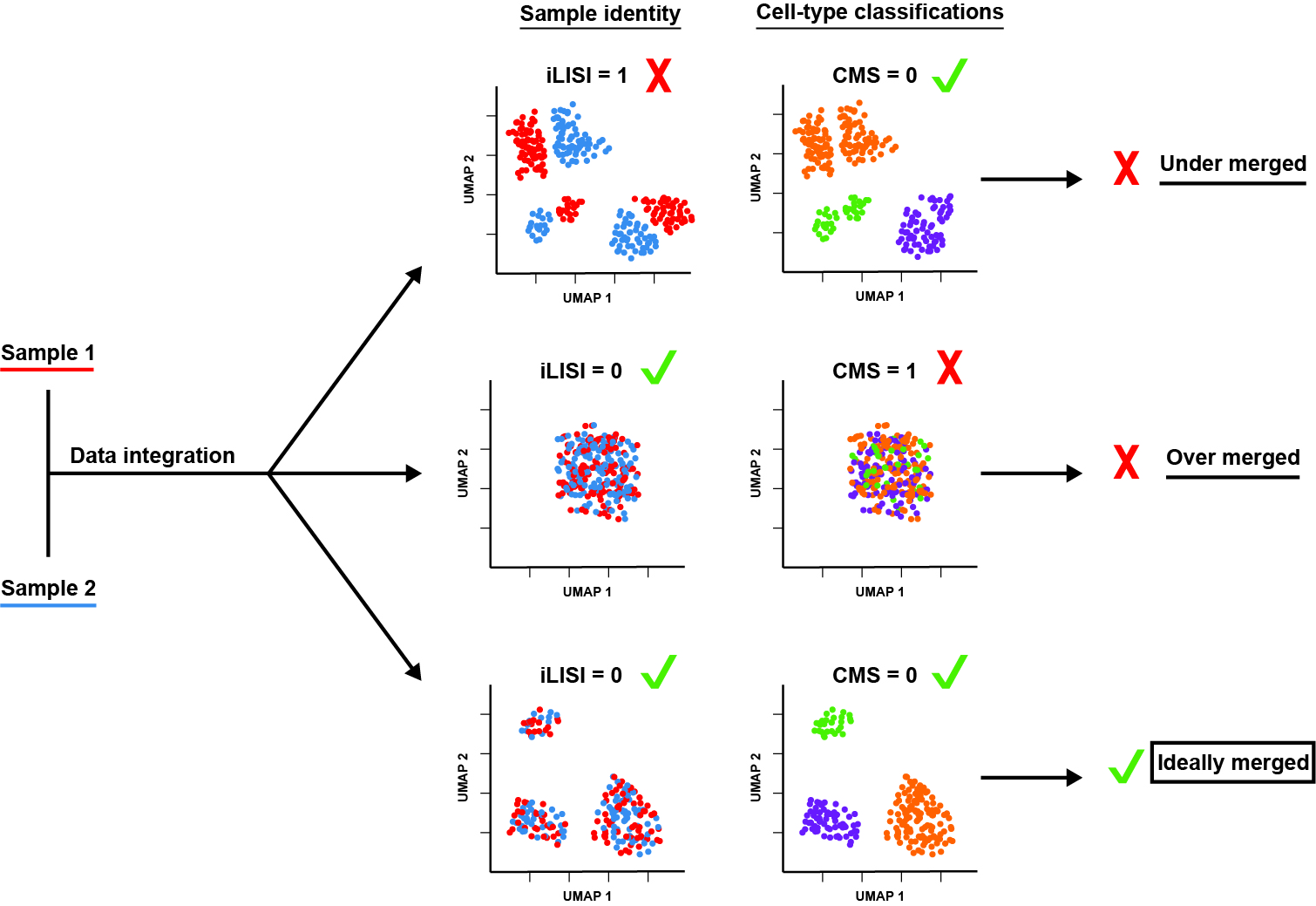
This repository is intended to support reproducibility of our recent manuscript: "Data Matrix Normalization and Merging Strategies Minimize Batch-specific Systemic Variation in scRNA-Seq Data." The complete text and supplementary materials of the preprint are made freely available via bioRxiv.[1]
To enable rapid reproduction of the published figures, we have made a set of pre-assembled Seurat objects available as part of an R packaged "BatchNorm", available on github here.
Installation
To install the package (along with select published datasets in support of the manuscript) in R, run:
if(!requireNamespace("devtools", quietly = TRUE)) {
install.packages("devtools")
}
devtools::install_github("Ghosn-Lab/BatchNorm")
The package should install within a few minutes, and all functions can be reproduced on a standard desktop or laptop without special hardware.
Examples
To enable reproducibility, we have published this repository as a user-friendly website here. Anyone with access to R can reproduce the figures in our manuscript by following along with our vignettes here
References
[1]
Babcock, B. R. et al.
Data Matrix Normalization and Merging Strategies Minimize Batch-specific Systemic Variation in scRNA-Seq Data.
bioRxiv (2021)
DOI: https://doi.org/10.1101/2021.08.18.456898
Ghosn-Lab/BatchNorm documentation built on Dec. 17, 2021, 9:32 p.m.
Ghosn Laboratory 
Data Matrix Normalization and Merging Strategies Minimize Batch-specific Systemic Variation in scRNA-Seq Data
This repository is intended to support reproducibility of our recent manuscript: "Data Matrix Normalization and Merging Strategies Minimize Batch-specific Systemic Variation in scRNA-Seq Data." The complete text and supplementary materials of the preprint are made freely available via bioRxiv.[1]
To enable rapid reproduction of the published figures, we have made a set of pre-assembled Seurat objects available as part of an R packaged "BatchNorm", available on github here.

Installation
To install the package (along with select published datasets in support of the manuscript) in R, run:
if(!requireNamespace("devtools", quietly = TRUE)) {
install.packages("devtools")
}
devtools::install_github("Ghosn-Lab/BatchNorm")
The package should install within a few minutes, and all functions can be reproduced on a standard desktop or laptop without special hardware.
Examples
To enable reproducibility, we have published this repository as a user-friendly website here. Anyone with access to R can reproduce the figures in our manuscript by following along with our vignettes here
References
[1] Babcock, B. R. et al. Data Matrix Normalization and Merging Strategies Minimize Batch-specific Systemic Variation in scRNA-Seq Data. bioRxiv (2021) DOI: https://doi.org/10.1101/2021.08.18.456898
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
