In GuangchuangYu/ggtreeghost:
ggtree ghost
just an idea of using ggghost to work with ggtree.
> tree
Phylogenetic tree with 30 tips and 29 internal nodes.
Tip labels:
t4, t18, t20, t10, t8, t12, ...
Rooted; includes branch lengths.
> p <- ggtreeghost(tree) + geom_hilight(40)
> code = summary(p, combine=TRUE)
> code
[1] "ggplot(tr, mapping = mapping, layout = layout, mrsd = mrsd, as.Date = as.Date, yscale = yscale, yscale_mapping = yscale_mapping, ladderize = ladderize, right = right, branch.length = branch.length, ndigits = ndigits) + geom_tree(layout = layout, multiPhylo = multiPhylo) + theme_tree() + geom_hilight(40)"
## reproduce the *plot* by running the code text
> eval(parse(text=code))
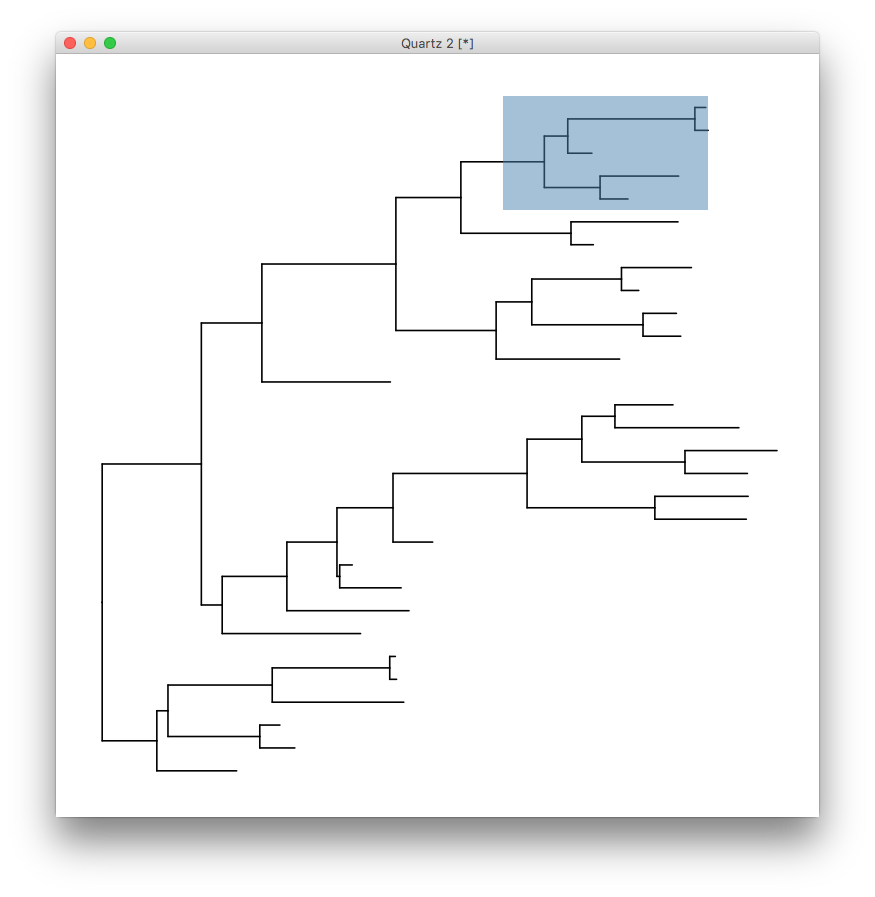
The tree will be stored in attr(p, "data")$data and with the
summary(p, combine=TRUE), we can output tree and source code to
visualize the tree (in GGTREE block) into a NEXSUS file.
Then by parsing the output NEXUS, we have the tree object and the
source code to reproduce tree visualization.
Just
similar to Figtree which can save the plot to NEXUS tree file. Figtree
introduce a figtree block in NEXUS file to store visualization
setting. The idea here is similar.
GuangchuangYu/ggtreeghost documentation built on May 6, 2019, 9:01 p.m.
ggtree ghost
just an idea of using ggghost to work with ggtree.
> tree Phylogenetic tree with 30 tips and 29 internal nodes. Tip labels: t4, t18, t20, t10, t8, t12, ... Rooted; includes branch lengths. > p <- ggtreeghost(tree) + geom_hilight(40) > code = summary(p, combine=TRUE) > code [1] "ggplot(tr, mapping = mapping, layout = layout, mrsd = mrsd, as.Date = as.Date, yscale = yscale, yscale_mapping = yscale_mapping, ladderize = ladderize, right = right, branch.length = branch.length, ndigits = ndigits) + geom_tree(layout = layout, multiPhylo = multiPhylo) + theme_tree() + geom_hilight(40)" ## reproduce the *plot* by running the code text > eval(parse(text=code))

The tree will be stored in attr(p, "data")$data and with the
summary(p, combine=TRUE), we can output tree and source code to
visualize the tree (in GGTREE block) into a NEXSUS file.
Then by parsing the output NEXUS, we have the tree object and the
source code to reproduce tree visualization.
Just
similar to Figtree which can save the plot to NEXUS tree file. Figtree
introduce a figtree block in NEXUS file to store visualization
setting. The idea here is similar.
Embedding an R snippet on your website
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
