In LieberInstitute/DeconvoBuddies: Helper Functions for LIBD Deconvolution
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
crop = NULL ## Related to https://stat.ethz.ch/pipermail/bioc-devel/2020-April/016656.html
)
## Bib setup
library("RefManageR")
## Write bibliography information
bib <- c(
R = citation(),
BiocStyle = citation("BiocStyle")[1],
knitr = citation("knitr")[1],
RefManageR = citation("RefManageR")[1],
rmarkdown = citation("rmarkdown")[1],
sessioninfo = citation("sessioninfo")[1],
testthat = citation("testthat")[1],
DeconvoBuddies = citation("DeconvoBuddies")[1]
)
Introduction
What are Marker Genes?
Cell type marker genes have cell type specific expression, that is high
expression in the target cell type, and low expression in all other cell
types. Sub-setting the genes considered in a cell type deconvolution
analysis helps reduce noise and can improve the accuracy of a
deconvolution method.
How can we select marker genes?
There are several approaches to select marker genes.
One popular method is "1 vs. All" differential expression [@lun], where
genes are tested for differential expression between the target cell
type, and a combined group of all "other" cell types. Statistically
significant differentially expressed genes (DEGs) can be selected as a
set of marker genes, DEGs can be ranked by high log fold change.
However in some cases 1vAll can select genes with high expression in
non-target cell types, especially in cell types related to the target
cell types (such as Neuron sub-types), or when there is a smaller number
of cells in the cell type and the signal is disguised within the other
group.
For example, in our snRNA-seq dataset from Human DLPFC
[@huuki-myers2024] selecting marker
gene for the cell type Oligodendrocyte (Oligo), MBP has a high log
fold change when testing by 1vALL (see illustration below). But, when
the expression of MBP is observed by individual cell types there is also
expression in the related cell types Microglia (Micro) and
Oligodendrocyte precursor cells (OPC).
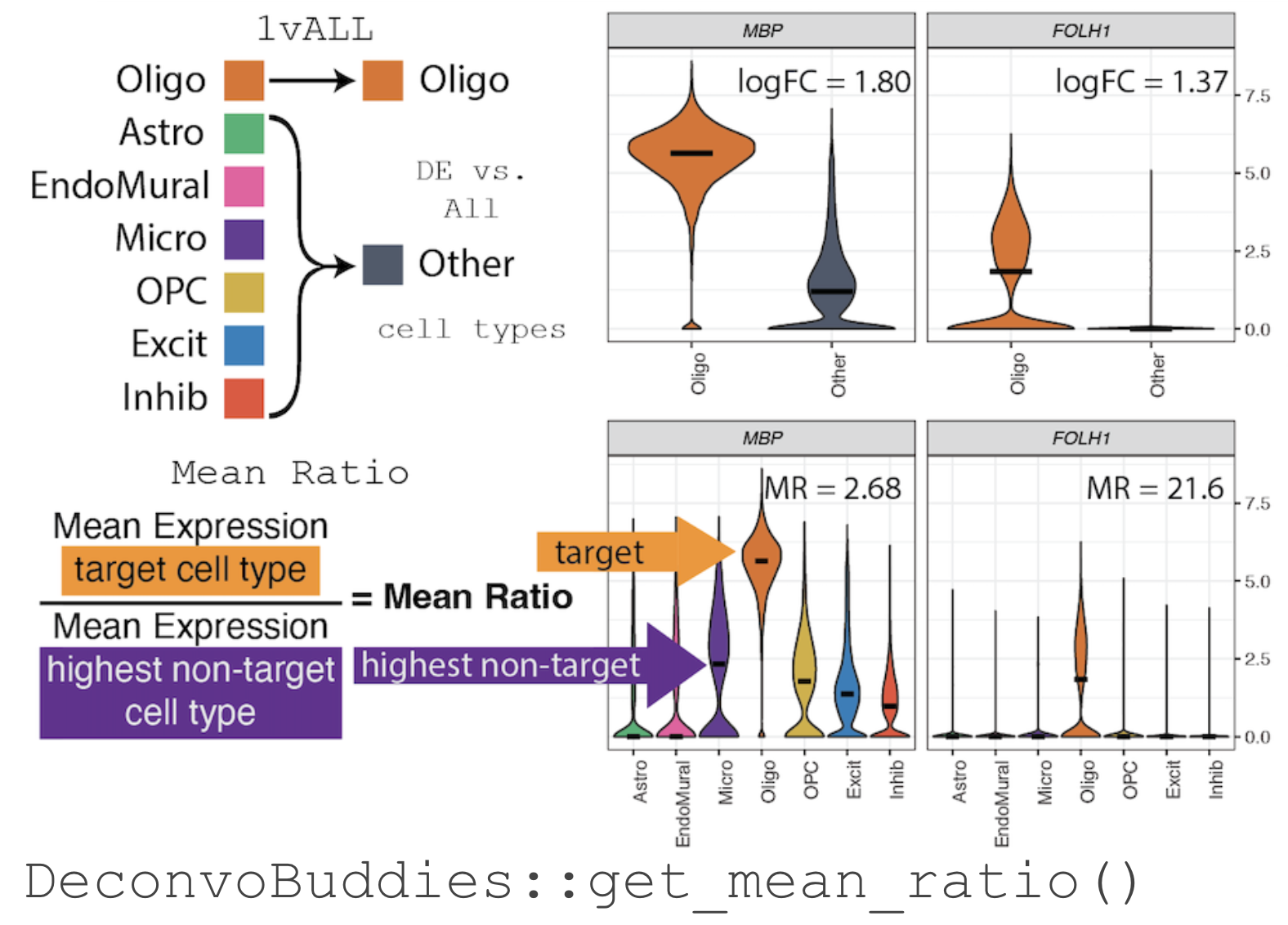
The Mean Ratio Method
To capture genes with more cell type specific expression and less noise,
we developed the Mean Ratio method. The Mean Ratio method works
by selecting genes with large differences between gene expression in the
target cell type and the closest non-target cell type, by evaluating
genes by their MeanRatio metric.
We calculate the MeanRatio for a target cell type for each gene by
dividing the mean expression of the target cell by the mean expression
of the next highest non-target cell type. Genes with the highest
MeanRatio values are selected as marker genes.
In the above example, Oligo is the target cell type. Micro has the
highest mean expression out of the other non-target (not Oligo) cell
types. The MeanRatio = (mean expression Oligo) / (mean expression Micro),
for MBP MeanRatio = 2.68 for gene FOLH1 MeanRatio is much higher
21.6 showing FOLH1 is the better marker gene (in contrast to ranking by
1vALL log FC). In the violin plots you can see that expression of
FOLH1 is much more specific to Oligo than MBP, supporting the ranking
by MeanRatio.
We have implemented the Mean Ratio method in this R package with
the function get_mean_ratio(). This vignette will cover our process for
marker gene selection.
Goals of this Vignette
We will be demonstrating how to use DeconvoBuddies tools when finding
cell type marker genes in single cell RNA-seq data via the MeanRatio
method.
- Install and load required packages
- Download DLPFC snRNA-seq data
- Find MeanRatio marker genes with
DeconvoBuddies::get_mean_ratio()
- Find 1vALL marker genes with
DeconvoBuddies::findMarkers_1vALL()
- Compare marker gene selection
- Visualize marker genes expression with
DeconcoBuddies::plot_gene_express() and related functions
1. Install and load required packages
R is an open-source statistical environment which can be easily
modified to enhance its functionality via packages.
r Biocpkg("DeconvoBuddies") is a R package available via the
Bioconductor repository for packages. R can
be installed on any operating system from
CRAN after which you can install
r Biocpkg("DeconvoBuddies") by using the following commands in your
R session:
Install DeconvoBuddies
if (!requireNamespace("BiocManager", quietly = TRUE)) {
install.packages("BiocManager")
}
BiocManager::install("DeconvoBuddies")
## Check that you have a valid Bioconductor installation
BiocManager::valid()
Load Other Packages
Let's load the packages will use in this vignette.
## Packages for different types of RNA-seq data structures in R
library("SingleCellExperiment")
## For downloading data
library("spatialLIBD")
## Other helper packages for this vignette
library("dplyr")
library("ggplot2")
## Our main package
library("DeconvoBuddies")
2. Download DLPFC snRNA-seq data.
Here we will download single nucleus RNA-seq data from the Human DLPFC
with 77k nuclei x 36k genes [@huuki-myers2024]. This data is stored in a
SingleCellExperiment object. The nuclei in this dataset are labeled by
cell types at a few resolutions, we will focus on the "broad" resolution
that contains seven cell types.
## Use spatialLIBD to fetch the snRNA-seq dataset
sce_path_zip <- fetch_deconvo_data("sce")
## unzip and load the data
sce_path <- unzip(sce_path_zip, exdir = tempdir())
sce <- HDF5Array::loadHDF5SummarizedExperiment(
file.path(tempdir(), "sce_DLPFC_annotated")
)
# lobstr::obj_size(sce)
# 172.28 MB
## exclude Ambiguous cell type
sce <- sce[, sce$cellType_broad_hc != "Ambiguous"]
sce$cellType_broad_hc <- droplevels(sce$cellType_broad_hc)
## Check the broad cell type distribution
table(sce$cellType_broad_hc)
## We're going to subset to the first 5k genes to save memory
## In a real application you'll want to use the full dataset
sce <- sce[seq_len(5000), ]
## check the final dimensions of the dataset
dim(sce)
3. Find MeanRatio marker genes
To find Mean Ratio marker genes for the data in sce we'll use the
function DeconvoBuddies::get_mean_ratio(), this function takes a
SingleCellExperiment object sce the name of the column in the
colData(sce) that contains the cell type annotations of interest (here
we'll use cellType_broad_hc), and optionally you can also supply
additional column names from the rowData(sce) to add the gene_name
and/or gene_ensembl information to the table output of
get_mean_ratio.
# calculate the Mean Ratio of genes for each cell type in sce
marker_stats_MeanRatio <- get_mean_ratio(
sce = sce, # sce is the SingleCellExperiment with our data
assay_name = "logcounts", ## assay to use, we recommend logcounts [default]
cellType_col = "cellType_broad_hc", # column in colData with cell type info
gene_ensembl = "gene_id", # column in rowData with ensembl gene ids
gene_name = "gene_name" # column in rowData with gene names/symbols
)
The function get_mean_ratio() returns a tibble with the following
columns:
gene is the name of the gene (from rownames(sce)).cellType.target is the cell type we're finding marker genes for.mean.target is the mean expression of gene for
cellType.target.cellType.2nd is the second highest non-target cell type.mean.2nd is the mean expression of gene for cellType.2nd.MeanRatio is the ratio of mean.target/mean.2nd.MeanRatio.rank is the rank of MeanRatio for the cell type.MeanRatio.anno is an annotation of the MeanRatio calculation
helpful for plotting.gene_ensembl & gene_name optional cols from rowData(sce)
specified by the user to add gene information
## Explore the tibble output
marker_stats_MeanRatio
## genes with the highest MeanRatio are the best marker genes for each cell type
marker_stats_MeanRatio |>
filter(MeanRatio.rank == 1)
4. Find 1vALL marker genes
To further explore cell type marker genes it can be helpful to also
calculate the 1vALL stats for the dataset. To help with this we have
included the function DeconvoBuddies::findMarkers_1vALL(), which is a
wrapper for scran::findMarkers() that iterates through cell types and
creates an table output in a compatible with the output
DeconvoBuddies::get_mean_ratio().
Similarity to get_mean_ratio this function requires the sce object,
cellType_col to define cell types, and assay_name. But
findMarkers_1vALL() also takes a model (mod) to use as design in
scran::findMarkers(), in this example we will control for donor which
is stored as BrNum.
Note this function can take a bit of time to run. We have saved the output as
data("marker_stats_1vAll") to speed-up the runtime of this vignette.
## Run 1vALL DE to find markers for each cell type - takes 5min+
# marker_stats_1vAll <- findMarkers_1vAll(
# sce = sce, # sce is the SingleCellExperiment with our data
# assay_name = "counts",
# cellType_col = "cellType_broad_hc", # column in colData with cell type info
# mod = "~BrNum" # Control for donor stored in "BrNum" with mod
# )
## load 1vAll data to save time, data is equivalent to the above code
data("marker_stats_1vAll")
The function findMarkers_1vALL() returns a tibble with the following
columns:
gene is the name of the gene (from rownames(sce)).logFC the log fold change from the DE testlog.p.value the log of the p-value of the DE testlog.FDR the log of the False Discovery Rate adjusted p.valuestd.logFC the standard logFCcellType.target the cell type we're finding marker genes forstd.logFC.rank the rank of std.logFC for each cell typestd.logFC.anno is an annotation of the std.logFC value helpful
for plotting.
## Explore the tibble output
marker_stats_1vAll
## genes with the highest MeanRatio are the best marker genes for each cell type
marker_stats_1vAll |>
filter(std.logFC.rank == 1)
As this is a differential expression test, you can create volcano plots
to explore the outputs. 🌋
Note that with the default option "up" for direction only up-regulated
genes are considered marker candidates, so all genes with logFC\<1 will
have a p.value=0. As a results these plots will only have the right half
of the volcano shape. If you'd like all p-values set
findMarkers_1vALL(direction="any").
# Create volcano plots of DE stats from 1vALL
marker_stats_1vAll |>
ggplot(aes(logFC, -log.p.value)) +
geom_point() +
facet_wrap(~cellType.target) +
geom_vline(xintercept = c(1, -1), linetype = "dashed", color = "red")
5. Compare Marker Gene Selection
Let's join the two marker_stats tables together to compare the
findings of the two methods.
Note as we are using a subset of data for this example, for some genes
there is not enough data to test and 1vALL will have some missing
results.
## join the two marker_stats tables
marker_stats <- marker_stats_MeanRatio |>
left_join(marker_stats_1vAll, by = join_by(gene, cellType.target))
## Check stats for our top genes
marker_stats |>
filter(MeanRatio.rank == 1) |>
select(gene, cellType.target, MeanRatio, MeanRatio.rank, std.logFC, std.logFC.rank)
Hockey Stick Plots
Plotting the values of Mean Ratio vs. standard log fold change (from
1vAll) we create what we call "hockey stick plots" 🏒. These plots help
visualize the distribution of MeanRatio and standard logFC values.
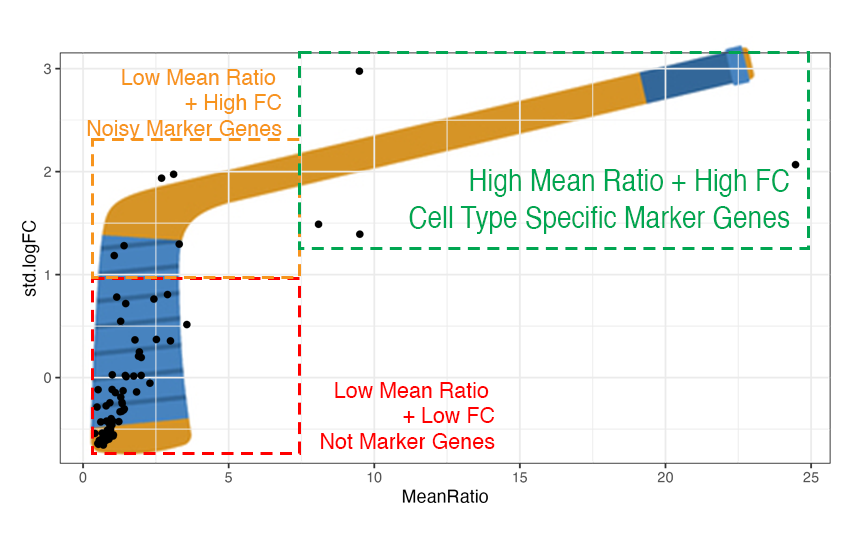
Typically for a cell type see most genes have low Mean Ratio and low
fold change, these genes are not marker genes (red box in illustration
above on the bottom left side).
Genes with higher fold change from 1vALL are better marker gene
candidates, but most have low MeanRatio values indicating that one or
more non-target cell types have high expression for that gene, causing
noise (orange box on the top left side).
Genes with high MeanRatio typically also have high 1vALL standard fold changes,
these are cell type specific marker genes we are selecting for (green
box on the top right side).
# create hockey stick plots to compare MeanRatio and standard logFC values.
marker_stats |>
ggplot(aes(MeanRatio, std.logFC)) +
geom_point() +
facet_wrap(~cellType.target)
We can see a "hockey stick" shape in most of the cell types, with a few
marker genes with high values for both logFC and MeanRatio.
6. Visualize Marker Genes Expression
An important step for ensuring you have selected high quality marker
genes is to visualize their expression over the cell types in the
dataset. DeconvoBuddies has several functions to help quickly plot
gene expression at a few levels:
plot_gene_express() plots the expression of one or more genes as a
violin plot.
## plot expression of two genes from a list
plot_gene_express(
sce = sce,
category = "cellType_broad_hc",
genes = c("SLC2A1", "CREG2")
)
plot_marker_express() plots the top n marker genes for a specified
cell type based on the values from marker_stats. Annotations for the
details of the MeanRatio value + calculation are added to each panel.
# plot the top 10 MeanRatio genes for Excit
plot_marker_express(
sce = sce,
stats = marker_stats,
cell_type = "Excit",
n_genes = 10,
cellType_col = "cellType_broad_hc"
)
In these violin plots we can see these genes have high expression in the
target cell type Excit , and mostly low expression nuclei in the other
cell types, sometimes even no expression.
This function defaults to selecting genes by the MeanRatio stats, but
can also be used to plot the 1vAll genes.
# plot the top 10 1vAll genes for Excit
plot_marker_express(
sce = sce,
stats = marker_stats,
cell_type = "Excit",
n_genes = 10,
rank_col = "std.logFC.rank", ## use logFC cols from 1vALL
anno_col = "std.logFC.anno",
cellType_col = "cellType_broad_hc"
)
We can see in the top 1vALL genes there is some expression of these
genes in Inhib nuclei in addition to the target cell type Excit.
plot_marker_express_ALL() plots the top marker genes for all cell
types. This is a quick and easy way to look at the the top markers in
your dataset, which is an important step and can help identify genes
with multimodal distributions that may confound the MeanRatio method.
# plot the top 10 1vAll genes for all cell types
print(plot_marker_express_ALL(
sce = sce,
stats = marker_stats,
n_genes = 10,
cellType_col = "cellType_broad_hc"
))
The violin plots can also be directly printed to a PDF file using the built
in argument plot_marker_express_ALL(pdf = "my_marker_genes.pdf") for
portability and easy sharing.
Summary
In this vignette we covered the importance of finding marker genes, and
introduced our method for finding cell type specific genes MeanRatio.
We covered how to find and compare MeanRatio marker genes with
get_mean_ratio(), and 1vALL marker genes with findMarkers_1vALL().
And finally, how to visualize the expression of these marker genes with
plot_marker_express() and related functions.
Next Steps
For an example of how to use these marker genes in a deconvolution
workflow, check out Vignette: Deconvolution Benchmark in Human
DLPFC.
We hope this vignette and DeconvoBuddies helps you with your research
goals! Thanks for reading 😁
Reproducibility
The r Biocpkg("DeconvoBuddies") package
r Citep(bib[["DeconvoBuddies"]]) was made possible thanks to:
- R
r Citep(bib[["R"]])
r Biocpkg("BiocStyle") r Citep(bib[["BiocStyle"]])r CRANpkg("knitr") r Citep(bib[["knitr"]])r CRANpkg("RefManageR") r Citep(bib[["RefManageR"]])r CRANpkg("rmarkdown") r Citep(bib[["rmarkdown"]])r CRANpkg("sessioninfo") r Citep(bib[["sessioninfo"]])r CRANpkg("testthat") r Citep(bib[["testthat"]])
This package was developed using r BiocStyle::Biocpkg("biocthis").
R session information.
## Session info
library("sessioninfo")
options(width = 120)
session_info()
Bibliography
This vignette was generated using r Biocpkg("BiocStyle")
r Citep(bib[["BiocStyle"]]) with r CRANpkg("knitr")
r Citep(bib[["knitr"]]) and r CRANpkg("rmarkdown")
r Citep(bib[["rmarkdown"]]) running behind the scenes.
Citations made with r CRANpkg("RefManageR")
r Citep(bib[["RefManageR"]]).
## Print bibliography
PrintBibliography(bib, .opts = list(hyperlink = "to.doc", style = "html"))
LieberInstitute/DeconvoBuddies documentation built on June 12, 2025, 11:03 a.m.
knitr::opts_chunk$set( collapse = TRUE, comment = "#>", crop = NULL ## Related to https://stat.ethz.ch/pipermail/bioc-devel/2020-April/016656.html )
## Bib setup library("RefManageR") ## Write bibliography information bib <- c( R = citation(), BiocStyle = citation("BiocStyle")[1], knitr = citation("knitr")[1], RefManageR = citation("RefManageR")[1], rmarkdown = citation("rmarkdown")[1], sessioninfo = citation("sessioninfo")[1], testthat = citation("testthat")[1], DeconvoBuddies = citation("DeconvoBuddies")[1] )
Introduction
What are Marker Genes?
Cell type marker genes have cell type specific expression, that is high expression in the target cell type, and low expression in all other cell types. Sub-setting the genes considered in a cell type deconvolution analysis helps reduce noise and can improve the accuracy of a deconvolution method.
How can we select marker genes?
There are several approaches to select marker genes.
One popular method is "1 vs. All" differential expression [@lun], where genes are tested for differential expression between the target cell type, and a combined group of all "other" cell types. Statistically significant differentially expressed genes (DEGs) can be selected as a set of marker genes, DEGs can be ranked by high log fold change.
However in some cases 1vAll can select genes with high expression in non-target cell types, especially in cell types related to the target cell types (such as Neuron sub-types), or when there is a smaller number of cells in the cell type and the signal is disguised within the other group.
For example, in our snRNA-seq dataset from Human DLPFC [@huuki-myers2024] selecting marker gene for the cell type Oligodendrocyte (Oligo), MBP has a high log fold change when testing by 1vALL (see illustration below). But, when the expression of MBP is observed by individual cell types there is also expression in the related cell types Microglia (Micro) and Oligodendrocyte precursor cells (OPC).

The Mean Ratio Method
To capture genes with more cell type specific expression and less noise,
we developed the Mean Ratio method. The Mean Ratio method works
by selecting genes with large differences between gene expression in the
target cell type and the closest non-target cell type, by evaluating
genes by their MeanRatio metric.
We calculate the MeanRatio for a target cell type for each gene by
dividing the mean expression of the target cell by the mean expression
of the next highest non-target cell type. Genes with the highest
MeanRatio values are selected as marker genes.
In the above example, Oligo is the target cell type. Micro has the
highest mean expression out of the other non-target (not Oligo) cell
types. The MeanRatio = (mean expression Oligo) / (mean expression Micro),
for MBP MeanRatio = 2.68 for gene FOLH1 MeanRatio is much higher
21.6 showing FOLH1 is the better marker gene (in contrast to ranking by
1vALL log FC). In the violin plots you can see that expression of
FOLH1 is much more specific to Oligo than MBP, supporting the ranking
by MeanRatio.
We have implemented the Mean Ratio method in this R package with
the function get_mean_ratio(). This vignette will cover our process for
marker gene selection.
Goals of this Vignette
We will be demonstrating how to use DeconvoBuddies tools when finding
cell type marker genes in single cell RNA-seq data via the MeanRatio
method.
- Install and load required packages
- Download DLPFC snRNA-seq data
- Find MeanRatio marker genes with
DeconvoBuddies::get_mean_ratio() - Find 1vALL marker genes with
DeconvoBuddies::findMarkers_1vALL() - Compare marker gene selection
- Visualize marker genes expression with
DeconcoBuddies::plot_gene_express()and related functions
1. Install and load required packages
R is an open-source statistical environment which can be easily
modified to enhance its functionality via packages.
r Biocpkg("DeconvoBuddies") is a R package available via the
Bioconductor repository for packages. R can
be installed on any operating system from
CRAN after which you can install
r Biocpkg("DeconvoBuddies") by using the following commands in your
R session:
Install DeconvoBuddies
if (!requireNamespace("BiocManager", quietly = TRUE)) { install.packages("BiocManager") } BiocManager::install("DeconvoBuddies") ## Check that you have a valid Bioconductor installation BiocManager::valid()
Load Other Packages
Let's load the packages will use in this vignette.
## Packages for different types of RNA-seq data structures in R library("SingleCellExperiment") ## For downloading data library("spatialLIBD") ## Other helper packages for this vignette library("dplyr") library("ggplot2") ## Our main package library("DeconvoBuddies")
2. Download DLPFC snRNA-seq data.
Here we will download single nucleus RNA-seq data from the Human DLPFC
with 77k nuclei x 36k genes [@huuki-myers2024]. This data is stored in a
SingleCellExperiment object. The nuclei in this dataset are labeled by
cell types at a few resolutions, we will focus on the "broad" resolution
that contains seven cell types.
## Use spatialLIBD to fetch the snRNA-seq dataset sce_path_zip <- fetch_deconvo_data("sce") ## unzip and load the data sce_path <- unzip(sce_path_zip, exdir = tempdir()) sce <- HDF5Array::loadHDF5SummarizedExperiment( file.path(tempdir(), "sce_DLPFC_annotated") ) # lobstr::obj_size(sce) # 172.28 MB ## exclude Ambiguous cell type sce <- sce[, sce$cellType_broad_hc != "Ambiguous"] sce$cellType_broad_hc <- droplevels(sce$cellType_broad_hc) ## Check the broad cell type distribution table(sce$cellType_broad_hc) ## We're going to subset to the first 5k genes to save memory ## In a real application you'll want to use the full dataset sce <- sce[seq_len(5000), ] ## check the final dimensions of the dataset dim(sce)
3. Find MeanRatio marker genes
To find Mean Ratio marker genes for the data in sce we'll use the
function DeconvoBuddies::get_mean_ratio(), this function takes a
SingleCellExperiment object sce the name of the column in the
colData(sce) that contains the cell type annotations of interest (here
we'll use cellType_broad_hc), and optionally you can also supply
additional column names from the rowData(sce) to add the gene_name
and/or gene_ensembl information to the table output of
get_mean_ratio.
# calculate the Mean Ratio of genes for each cell type in sce marker_stats_MeanRatio <- get_mean_ratio( sce = sce, # sce is the SingleCellExperiment with our data assay_name = "logcounts", ## assay to use, we recommend logcounts [default] cellType_col = "cellType_broad_hc", # column in colData with cell type info gene_ensembl = "gene_id", # column in rowData with ensembl gene ids gene_name = "gene_name" # column in rowData with gene names/symbols )
The function get_mean_ratio() returns a tibble with the following
columns:
geneis the name of the gene (from rownames(sce)).cellType.targetis the cell type we're finding marker genes for.mean.targetis the mean expression ofgeneforcellType.target.cellType.2ndis the second highest non-target cell type.mean.2ndis the mean expression ofgeneforcellType.2nd.MeanRatiois the ratio ofmean.target/mean.2nd.MeanRatio.rankis the rank ofMeanRatiofor the cell type.MeanRatio.annois an annotation of theMeanRatiocalculation helpful for plotting.gene_ensembl&gene_nameoptional cols fromrowData(sce)specified by the user to add gene information
## Explore the tibble output marker_stats_MeanRatio ## genes with the highest MeanRatio are the best marker genes for each cell type marker_stats_MeanRatio |> filter(MeanRatio.rank == 1)
4. Find 1vALL marker genes
To further explore cell type marker genes it can be helpful to also
calculate the 1vALL stats for the dataset. To help with this we have
included the function DeconvoBuddies::findMarkers_1vALL(), which is a
wrapper for scran::findMarkers() that iterates through cell types and
creates an table output in a compatible with the output
DeconvoBuddies::get_mean_ratio().
Similarity to get_mean_ratio this function requires the sce object,
cellType_col to define cell types, and assay_name. But
findMarkers_1vALL() also takes a model (mod) to use as design in
scran::findMarkers(), in this example we will control for donor which
is stored as BrNum.
Note this function can take a bit of time to run. We have saved the output as
data("marker_stats_1vAll") to speed-up the runtime of this vignette.
## Run 1vALL DE to find markers for each cell type - takes 5min+ # marker_stats_1vAll <- findMarkers_1vAll( # sce = sce, # sce is the SingleCellExperiment with our data # assay_name = "counts", # cellType_col = "cellType_broad_hc", # column in colData with cell type info # mod = "~BrNum" # Control for donor stored in "BrNum" with mod # ) ## load 1vAll data to save time, data is equivalent to the above code data("marker_stats_1vAll")
The function findMarkers_1vALL() returns a tibble with the following
columns:
geneis the name of the gene (from rownames(sce)).logFCthe log fold change from the DE testlog.p.valuethe log of the p-value of the DE testlog.FDRthe log of the False Discovery Rate adjusted p.valuestd.logFCthe standard logFCcellType.targetthe cell type we're finding marker genes forstd.logFC.rankthe rank ofstd.logFCfor each cell typestd.logFC.annois an annotation of thestd.logFCvalue helpful for plotting.
## Explore the tibble output marker_stats_1vAll ## genes with the highest MeanRatio are the best marker genes for each cell type marker_stats_1vAll |> filter(std.logFC.rank == 1)
As this is a differential expression test, you can create volcano plots to explore the outputs. 🌋
Note that with the default option "up" for direction only up-regulated
genes are considered marker candidates, so all genes with logFC\<1 will
have a p.value=0. As a results these plots will only have the right half
of the volcano shape. If you'd like all p-values set
findMarkers_1vALL(direction="any").
# Create volcano plots of DE stats from 1vALL marker_stats_1vAll |> ggplot(aes(logFC, -log.p.value)) + geom_point() + facet_wrap(~cellType.target) + geom_vline(xintercept = c(1, -1), linetype = "dashed", color = "red")
5. Compare Marker Gene Selection
Let's join the two marker_stats tables together to compare the
findings of the two methods.
Note as we are using a subset of data for this example, for some genes there is not enough data to test and 1vALL will have some missing results.
## join the two marker_stats tables marker_stats <- marker_stats_MeanRatio |> left_join(marker_stats_1vAll, by = join_by(gene, cellType.target)) ## Check stats for our top genes marker_stats |> filter(MeanRatio.rank == 1) |> select(gene, cellType.target, MeanRatio, MeanRatio.rank, std.logFC, std.logFC.rank)
Hockey Stick Plots
Plotting the values of Mean Ratio vs. standard log fold change (from
1vAll) we create what we call "hockey stick plots" 🏒. These plots help
visualize the distribution of MeanRatio and standard logFC values.

Typically for a cell type see most genes have low Mean Ratio and low fold change, these genes are not marker genes (red box in illustration above on the bottom left side).
Genes with higher fold change from 1vALL are better marker gene candidates, but most have low MeanRatio values indicating that one or more non-target cell types have high expression for that gene, causing noise (orange box on the top left side).
Genes with high MeanRatio typically also have high 1vALL standard fold changes, these are cell type specific marker genes we are selecting for (green box on the top right side).
# create hockey stick plots to compare MeanRatio and standard logFC values. marker_stats |> ggplot(aes(MeanRatio, std.logFC)) + geom_point() + facet_wrap(~cellType.target)
We can see a "hockey stick" shape in most of the cell types, with a few
marker genes with high values for both logFC and MeanRatio.
6. Visualize Marker Genes Expression
An important step for ensuring you have selected high quality marker
genes is to visualize their expression over the cell types in the
dataset. DeconvoBuddies has several functions to help quickly plot
gene expression at a few levels:
plot_gene_express() plots the expression of one or more genes as a
violin plot.
## plot expression of two genes from a list plot_gene_express( sce = sce, category = "cellType_broad_hc", genes = c("SLC2A1", "CREG2") )
plot_marker_express() plots the top n marker genes for a specified
cell type based on the values from marker_stats. Annotations for the
details of the MeanRatio value + calculation are added to each panel.
# plot the top 10 MeanRatio genes for Excit plot_marker_express( sce = sce, stats = marker_stats, cell_type = "Excit", n_genes = 10, cellType_col = "cellType_broad_hc" )
In these violin plots we can see these genes have high expression in the target cell type Excit , and mostly low expression nuclei in the other cell types, sometimes even no expression.
This function defaults to selecting genes by the MeanRatio stats, but can also be used to plot the 1vAll genes.
# plot the top 10 1vAll genes for Excit plot_marker_express( sce = sce, stats = marker_stats, cell_type = "Excit", n_genes = 10, rank_col = "std.logFC.rank", ## use logFC cols from 1vALL anno_col = "std.logFC.anno", cellType_col = "cellType_broad_hc" )
We can see in the top 1vALL genes there is some expression of these genes in Inhib nuclei in addition to the target cell type Excit.
plot_marker_express_ALL() plots the top marker genes for all cell
types. This is a quick and easy way to look at the the top markers in
your dataset, which is an important step and can help identify genes
with multimodal distributions that may confound the MeanRatio method.
# plot the top 10 1vAll genes for all cell types print(plot_marker_express_ALL( sce = sce, stats = marker_stats, n_genes = 10, cellType_col = "cellType_broad_hc" ))
The violin plots can also be directly printed to a PDF file using the built
in argument plot_marker_express_ALL(pdf = "my_marker_genes.pdf") for
portability and easy sharing.
Summary
In this vignette we covered the importance of finding marker genes, and
introduced our method for finding cell type specific genes MeanRatio.
We covered how to find and compare MeanRatio marker genes with
get_mean_ratio(), and 1vALL marker genes with findMarkers_1vALL().
And finally, how to visualize the expression of these marker genes with
plot_marker_express() and related functions.
Next Steps
For an example of how to use these marker genes in a deconvolution workflow, check out Vignette: Deconvolution Benchmark in Human DLPFC.
We hope this vignette and DeconvoBuddies helps you with your research goals! Thanks for reading 😁
Reproducibility
The r Biocpkg("DeconvoBuddies") package
r Citep(bib[["DeconvoBuddies"]]) was made possible thanks to:
- R
r Citep(bib[["R"]]) r Biocpkg("BiocStyle")r Citep(bib[["BiocStyle"]])r CRANpkg("knitr")r Citep(bib[["knitr"]])r CRANpkg("RefManageR")r Citep(bib[["RefManageR"]])r CRANpkg("rmarkdown")r Citep(bib[["rmarkdown"]])r CRANpkg("sessioninfo")r Citep(bib[["sessioninfo"]])r CRANpkg("testthat")r Citep(bib[["testthat"]])
This package was developed using r BiocStyle::Biocpkg("biocthis").
R session information.
## Session info library("sessioninfo") options(width = 120) session_info()
Bibliography
This vignette was generated using r Biocpkg("BiocStyle")
r Citep(bib[["BiocStyle"]]) with r CRANpkg("knitr")
r Citep(bib[["knitr"]]) and r CRANpkg("rmarkdown")
r Citep(bib[["rmarkdown"]]) running behind the scenes.
Citations made with r CRANpkg("RefManageR")
r Citep(bib[["RefManageR"]]).
## Print bibliography PrintBibliography(bib, .opts = list(hyperlink = "to.doc", style = "html"))
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
