In Miachol/BioInstaller: Integrator of Bioinformatics Resources
knitr::opts_chunk$set(echo = TRUE, screenshot.force = FALSE, comment = "#>", collapse = TRUE)
Introduction
Shiny application of BioInstaller has been supported since v0.3.5. A pre-built Docker image can help you to start all configured BioInstaller services in one-click, which includes the R (v3.4.4), Opencpu server, Shiny server, and latest BioInstaller.
How to deploy the BioInstaller using Docker
You need install the Docker program for futher opreations. It is required for running any Docker image including BioInstaller.
```{bash eval=FALSE}
docker pull bioinstaller/bioinstaller
docker run -it -p 80:80 -p 8004:8004 -v /tmp/download:/tmp/download bioinstaller/bioinstaller
When the docker container of BioInstaller was created, you can access the three types of web service in the browser.
- localhost/ocpu/ Opencpu server
- localhost/shiny/BioInstaller Shiny server
- localhost/rstudio/ Rstudio server (opencpu/opencpu)
### Opencpu server

```bash
#Show all items supported by BioInstaller
curl http://localhost/ocpu/library/BioInstaller/R/install.bioinfo -d \
"show.all.names=TRUE" X POST
#Show all versions of Bwa
curl http://localhost/ocpu/library/BioInstaller/R/install.bioinfo \
-d "name='bwa', show.all.versions=TRUE" -X POST
#Download and install latest Bwa
curl http://localhost/ocpu/library/BioInstaller/R/install.bioinfo \
-d "name='bwa', destdir='/opt/aliner/bwa'" -X POST
# Download 1000 Genome Project annotation database
curl http://localhost:5656/ocpu/library/BioInstaller/R/install.bioinfo \
-d "name='db_annovar_1000g', destdir = '/opt/annovardb', \
extra.list=list(buildver='hg19')" -X POST
# Get the character key and retrieve the output
# Get the JSON format value of returned output
curl http://localhost/ocpu/tmp/{key}/R/.val/json
# Get the text format value of returned output
curl http://localhost/ocpu/tmp/{key}/R/.val/text
# Get the function ‘print’ output of returned value
curl http://localhost/ocpu/tmp/{key}/R/.val/print
Shiny server
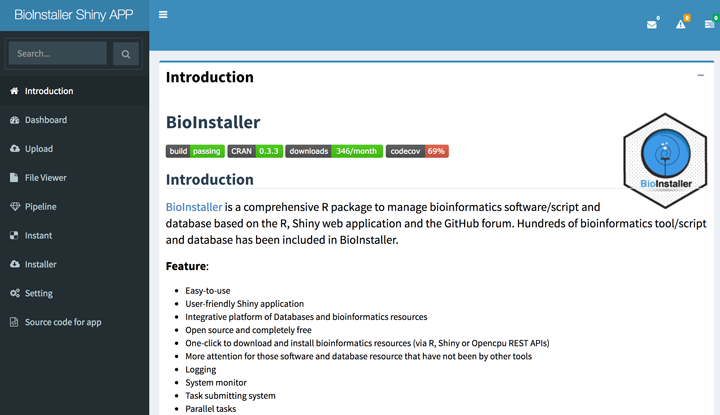
Open browser and input the URL "localhost/shiny/BioInstaller". It will activate a Shiny session and start the BioInstaller Shiny application.
Rstudio server
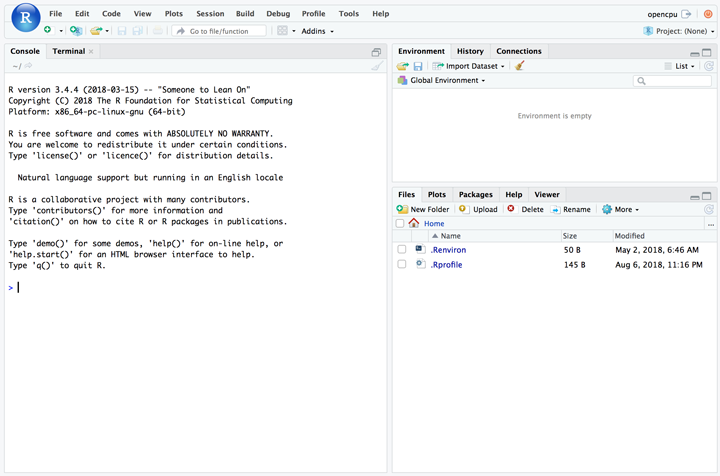
The Rstudio server was provided for users to connect the docker container just like in the desktop version of Rstudio.
How to start the standalone Shiny service of bioshiny
If the users can not to run a docker image, the standalone shiny service of bioshiny is an another way to start the service.
echo 'export BIO_SOFTWARES_DB_ACTIVE="~/.bioshiny/info.yaml" >> ~/.bashrc'
echo 'export BIOSHINY_CONFIG="~/.bioshiny/shiny.config.yaml" >> ~/.bashrc'
. ~/.bashrc
# Start the standalone Shiny application
wget https://raw.githubusercontent.com/openbiox/bioshiny/master/bin/bioshiny_deps_r
wget https://raw.githubusercontent.com/openbiox/bioshiny/master/bin/bioshiny_start
chmod a+x bioshiny_deps_r
chmod a+x bioshiny_start
./bioshiny_deps_r
# Start Shiny application workers
Rscript -e "bioshiny::set_shiny_workers(1)"
./bioshiny_start
# or use yarn
yarn global add bioshiny
bioshiny_deps_r
Rscript -e "bioshiny::set_shiny_workers(1)"
bioshiny_start
# If the bioshiny R package has not been installed,
# you need execute the followed commands:
# devtools::install_github('openbiox/bioshiny/src/bioshiny')
# It is better to start the worker in another background processor
bioshiny::set_shiny_workers(3)
bioshiny::web(auto_create = TRUE)
Miachol/BioInstaller documentation built on Feb. 1, 2023, 8:11 a.m.
knitr::opts_chunk$set(echo = TRUE, screenshot.force = FALSE, comment = "#>", collapse = TRUE)
Introduction
Shiny application of BioInstaller has been supported since v0.3.5. A pre-built Docker image can help you to start all configured BioInstaller services in one-click, which includes the R (v3.4.4), Opencpu server, Shiny server, and latest BioInstaller.
How to deploy the BioInstaller using Docker
You need install the Docker program for futher opreations. It is required for running any Docker image including BioInstaller.
```{bash eval=FALSE} docker pull bioinstaller/bioinstaller docker run -it -p 80:80 -p 8004:8004 -v /tmp/download:/tmp/download bioinstaller/bioinstaller
When the docker container of BioInstaller was created, you can access the three types of web service in the browser.
- localhost/ocpu/ Opencpu server
- localhost/shiny/BioInstaller Shiny server
- localhost/rstudio/ Rstudio server (opencpu/opencpu)
### Opencpu server

```bash
#Show all items supported by BioInstaller
curl http://localhost/ocpu/library/BioInstaller/R/install.bioinfo -d \
"show.all.names=TRUE" X POST
#Show all versions of Bwa
curl http://localhost/ocpu/library/BioInstaller/R/install.bioinfo \
-d "name='bwa', show.all.versions=TRUE" -X POST
#Download and install latest Bwa
curl http://localhost/ocpu/library/BioInstaller/R/install.bioinfo \
-d "name='bwa', destdir='/opt/aliner/bwa'" -X POST
# Download 1000 Genome Project annotation database
curl http://localhost:5656/ocpu/library/BioInstaller/R/install.bioinfo \
-d "name='db_annovar_1000g', destdir = '/opt/annovardb', \
extra.list=list(buildver='hg19')" -X POST
# Get the character key and retrieve the output
# Get the JSON format value of returned output
curl http://localhost/ocpu/tmp/{key}/R/.val/json
# Get the text format value of returned output
curl http://localhost/ocpu/tmp/{key}/R/.val/text
# Get the function ‘print’ output of returned value
curl http://localhost/ocpu/tmp/{key}/R/.val/print
Shiny server
Open browser and input the URL "localhost/shiny/BioInstaller". It will activate a Shiny session and start the BioInstaller Shiny application.

Rstudio server
The Rstudio server was provided for users to connect the docker container just like in the desktop version of Rstudio.

How to start the standalone Shiny service of bioshiny
If the users can not to run a docker image, the standalone shiny service of bioshiny is an another way to start the service.
echo 'export BIO_SOFTWARES_DB_ACTIVE="~/.bioshiny/info.yaml" >> ~/.bashrc' echo 'export BIOSHINY_CONFIG="~/.bioshiny/shiny.config.yaml" >> ~/.bashrc' . ~/.bashrc # Start the standalone Shiny application wget https://raw.githubusercontent.com/openbiox/bioshiny/master/bin/bioshiny_deps_r wget https://raw.githubusercontent.com/openbiox/bioshiny/master/bin/bioshiny_start chmod a+x bioshiny_deps_r chmod a+x bioshiny_start ./bioshiny_deps_r # Start Shiny application workers Rscript -e "bioshiny::set_shiny_workers(1)" ./bioshiny_start # or use yarn yarn global add bioshiny bioshiny_deps_r Rscript -e "bioshiny::set_shiny_workers(1)" bioshiny_start
# If the bioshiny R package has not been installed, # you need execute the followed commands: # devtools::install_github('openbiox/bioshiny/src/bioshiny') # It is better to start the worker in another background processor bioshiny::set_shiny_workers(3) bioshiny::web(auto_create = TRUE)
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
