README.md
In PatrickLeBlanc/LTNLDA: An implementation of a collapsed blocked Gibbs Sampler for the LTN-LDA model
LTNLDA
LTNLDA is a package which implements the LTN-LDA model. LTN-LDA (LeBlanc
and Ma XXXX) is a mixed-membership model which seeks to appropriately
incorporate cross-sample heterogeneity in subcommunity compositions: a
characteristic of the data prevalent in most microbiome studies.
Incorporating such cross-sample heterogeneity leads to substantially
improved inference compared to existing models.
Installation
You can install the LTNLDA from GitHub with the
following code:
# install.packages("devtools")
devtools::install_github("PatrickLeBlanc/LTNLDA", build_vignettes = TRUE)
Functions
There are two main functions included in the LTNLDA package. The first
is LTNLDA, which implements a collapsed blocked Gibbs sampler for the
LTN-LDA model. The second is LTNLDA_Perplexity, which finds the
perplexity of a fitted LTN-LDA model on a test set.
LTNLDA
We present code for running LTNLDA on the dataset included in the LTNLDA
package, a modified version of the data collected in (Dethlefsen and
Relman 2011). The user must choose the number of subcommunities and the
threshold controlling cross-sample heterogeneity. The following code is
presented, but not run. A more detailed explanation is presented in the
“LTN-LDA” vignette.
library(LTNLDA)
#load the data included with the package
data("ps",package = "LTNLDA")
#choose the number of subcommunities
K = 2
#fit the model
model = LTNLDA(ps,K)
Summary
Summary is a function which provides a high level summary of the model’s
inference. We include the average abundance of subcommunities across
samples as well as the top
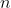 ASVs in each subcommunity and their prevalances. More information is
included in the “LTN-LDA” vignette.
ASVs in each subcommunity and their prevalances. More information is
included in the “LTN-LDA” vignette.
sum = Summary(model)
LTNLDA_Perplexity
We present code for running LTNLDA_perplexity on the dataset included
in the LTNLDA package, a modified version of the data collected in
(Dethlefsen and Relman 2011). The user first fits the LTN-LDA model on a
training set using the LTNLDA function featured in the previous section.
Then, the user uses the LTNLDA_Perplexity function to evaluate the
perplexity of the fitted model on the test set. The following code is
presented, but not run. A more detailed explanation is presented in the
“Perplexity” vignette.
library(LTNLDA)
set.seed(1)
#load dataset
data("ps",package = "LTNLDA")
#find total number of samples
num_samples = ncol(otu_table(ps))
#find the number of samples in the test set if we partition our dataset in half
num_test_samples = round(num_samples/2)
#randomly determine which samples are in the test set
test_samples = sample(1:num_samples,num_test_samples)
#make a vector such that the d^th entry denotes the set membership of sample d
set = rep("Train",num_samples)
set[test_samples] = "Test"
#Add this vector to the sample data of the phyloseq object
metadata = sample_data(ps)
metadata$Set = set
sample_data(ps) = metadata
#Partition the ps object into training and test sets
train_ps = subset_samples(ps, set == "Train")
test_ps = subset_samples(ps, set == "Test")
#Run Analysis
#choose the number of subcommunities
K = 2
#fit the model
model = LTNLDA(ps,K)
#run perplexity analysis analysis
perp = LTNLDA_Perplexity(model = model, ps = test_ps)
block_LTNLDA
This function runs a Gibbs sampler for the Block LTN-LDA model. We
recommend using the LTN-LDA model instead of the block LTN-LDA model,
but provide the user with the tools to run this model if they so wish.
Vignettes
To access the vignettes, use this code:
browseVignettes("LTNLDA")
References
- Les Dethlefsen and David A. Relman. Incomplete recovery and
individualized responses of the human distal gut microbiota to
repeated antibiotic perturbation. Proceedings of the National
Academy of the Sciences of the United States of America.
18(Supplement 1): 4554-4561, 2011.
- Patrick LeBlanc and Li Ma. Microbiome subcommunity learning with
logistic-tree normal latent Dirichlet allocation.
https://arxiv.org/abs/2109.05386, 2022.
Additionally, I inspiration for coding a collapsed LDA Gibbs sampler
from:
- Brooks, Andrew. “Latent Dirichlet Allocation – under the hood”. Web
blog post. data science side projects, thoughts, & experiments.
github.io. January 17, 2015. Web. September 11, 2021.
PatrickLeBlanc/LTNLDA documentation built on May 22, 2022, 12:49 p.m.
LTNLDA
LTNLDA is a package which implements the LTN-LDA model. LTN-LDA (LeBlanc and Ma XXXX) is a mixed-membership model which seeks to appropriately incorporate cross-sample heterogeneity in subcommunity compositions: a characteristic of the data prevalent in most microbiome studies. Incorporating such cross-sample heterogeneity leads to substantially improved inference compared to existing models.
Installation
You can install the LTNLDA from GitHub with the following code:
# install.packages("devtools")
devtools::install_github("PatrickLeBlanc/LTNLDA", build_vignettes = TRUE)
Functions
There are two main functions included in the LTNLDA package. The first is LTNLDA, which implements a collapsed blocked Gibbs sampler for the LTN-LDA model. The second is LTNLDA_Perplexity, which finds the perplexity of a fitted LTN-LDA model on a test set.
LTNLDA
We present code for running LTNLDA on the dataset included in the LTNLDA package, a modified version of the data collected in (Dethlefsen and Relman 2011). The user must choose the number of subcommunities and the threshold controlling cross-sample heterogeneity. The following code is presented, but not run. A more detailed explanation is presented in the “LTN-LDA” vignette.
library(LTNLDA)
#load the data included with the package
data("ps",package = "LTNLDA")
#choose the number of subcommunities
K = 2
#fit the model
model = LTNLDA(ps,K)
Summary
Summary is a function which provides a high level summary of the model’s
inference. We include the average abundance of subcommunities across
samples as well as the top
ASVs in each subcommunity and their prevalances. More information is
included in the “LTN-LDA” vignette.
sum = Summary(model)
LTNLDA_Perplexity
We present code for running LTNLDA_perplexity on the dataset included in the LTNLDA package, a modified version of the data collected in (Dethlefsen and Relman 2011). The user first fits the LTN-LDA model on a training set using the LTNLDA function featured in the previous section. Then, the user uses the LTNLDA_Perplexity function to evaluate the perplexity of the fitted model on the test set. The following code is presented, but not run. A more detailed explanation is presented in the “Perplexity” vignette.
library(LTNLDA)
set.seed(1)
#load dataset
data("ps",package = "LTNLDA")
#find total number of samples
num_samples = ncol(otu_table(ps))
#find the number of samples in the test set if we partition our dataset in half
num_test_samples = round(num_samples/2)
#randomly determine which samples are in the test set
test_samples = sample(1:num_samples,num_test_samples)
#make a vector such that the d^th entry denotes the set membership of sample d
set = rep("Train",num_samples)
set[test_samples] = "Test"
#Add this vector to the sample data of the phyloseq object
metadata = sample_data(ps)
metadata$Set = set
sample_data(ps) = metadata
#Partition the ps object into training and test sets
train_ps = subset_samples(ps, set == "Train")
test_ps = subset_samples(ps, set == "Test")
#Run Analysis
#choose the number of subcommunities
K = 2
#fit the model
model = LTNLDA(ps,K)
#run perplexity analysis analysis
perp = LTNLDA_Perplexity(model = model, ps = test_ps)
block_LTNLDA
This function runs a Gibbs sampler for the Block LTN-LDA model. We recommend using the LTN-LDA model instead of the block LTN-LDA model, but provide the user with the tools to run this model if they so wish.
Vignettes
To access the vignettes, use this code:
browseVignettes("LTNLDA")
References
- Les Dethlefsen and David A. Relman. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proceedings of the National Academy of the Sciences of the United States of America. 18(Supplement 1): 4554-4561, 2011.
- Patrick LeBlanc and Li Ma. Microbiome subcommunity learning with logistic-tree normal latent Dirichlet allocation. https://arxiv.org/abs/2109.05386, 2022.
Additionally, I inspiration for coding a collapsed LDA Gibbs sampler from:
- Brooks, Andrew. “Latent Dirichlet Allocation – under the hood”. Web blog post. data science side projects, thoughts, & experiments. github.io. January 17, 2015. Web. September 11, 2021.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
