README.md
In TMasak/surfcov: Random Surfaces' Covariance Estimation beyond Separability
surfcov
The purpose of R package surfcov is to enable covariance estimation
for random surfaces beyond separability, proposed in the papers
arXiv:1912.12870 and
arXiv:2007.12175.
Let
 be i.i.d. matrices of size
be i.i.d. matrices of size
 representing discrete measurements (on a grid) of some latent random
surfaces on a 2D domain, and
representing discrete measurements (on a grid) of some latent random
surfaces on a 2D domain, and
 be the covariance operator. The covariance is a tensor of size
be the covariance operator. The covariance is a tensor of size
 ,
which becomes problematic to handle for
,
which becomes problematic to handle for
 and
and
 as small as 100. The assumption of separability postulates that
as small as 100. The assumption of separability postulates that
![C[i,j,i',j'] = C_1[i,i'] C_2[j,j'],](https://latex.codecogs.com/png.image?%5Cdpi%7B110%7D&space;%5Cbg_white&space;C%5Bi%2Cj%2Ci%27%2Cj%27%5D%20%3D%20C_1%5Bi%2Ci%27%5D%20C_2%5Bj%2Cj%27%5D%2C)
which reduces statistical and computational burden, but is often
critized as an oversimplification, since it does not allow any
interaction between the two dimensions.
This package allows for efficient estimation and subsequent manipulation
of two alternative models, which are both strict generalizations of
separability:
- the separable-plus-banded model:
![[i,j,i',j'] = A_1[i,i'] A_2[j,j'] + B[i,j,i',j']](https://latex.codecogs.com/png.image?%5Cdpi%7B110%7D&space;%5Cbg_white&space;%5Bi%2Cj%2Ci%27%2Cj%27%5D%20%3D%20A_1%5Bi%2Ci%27%5D%20A_2%5Bj%2Cj%27%5D%20%2B%20B%5Bi%2Cj%2Ci%27%2Cj%27%5D) ,
where
,
where
![B[i,j,i',j'] = 0](https://latex.codecogs.com/png.image?%5Cdpi%7B110%7D&space;%5Cbg_white&space;B%5Bi%2Cj%2Ci%27%2Cj%27%5D%20%3D%200) for
for
 or
or
 ;
;
- the
 -separable
model:
-separable
model:
![C[i,j,i',j'] = A_1[i,i']B_1[j,j'] + \ldots + A_R[i,i']B_R[j,j']](https://latex.codecogs.com/png.image?%5Cdpi%7B110%7D&space;%5Cbg_white&space;C%5Bi%2Cj%2Ci%27%2Cj%27%5D%20%3D%20A_1%5Bi%2Ci%27%5DB_1%5Bj%2Cj%27%5D%20%2B%20%5Cldots%20%2B%20A_R%5Bi%2Ci%27%5DB_R%5Bj%2Cj%27%5D) .
.
When the data are stored in form of an array X of size
 ,
it can be simply checked whether a given model can be potentially useful
by running
,
it can be simply checked whether a given model can be potentially useful
by running
spb(X) for the separable-plus-banded model; orscd(X) for the
 -separable
model.
-separable
model.
Installation
You can install the development version from
GitHub with:
install.packages("devtools") # only if devtools is not yet installed
library(devtools)
install_github("TMasak/surfcov")
Examples
When the data are stored in form of an array X of size
 ,
it can be simply checked whether a given model can be potentially useful
by running
,
it can be simply checked whether a given model can be potentially useful
by running
spb(X) for the separable-plus-banded model; orscd(X) for the
 -separable
model.
-separable
model.
In particular,
# X <- array(runif(100*20*30),c(100,20,30))
spb_est <- spb(X)
spb$d
Run cross-validation (CV) in order to pick a value of the bandwidth
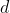 ,
and then fits the model with the best
,
and then fits the model with the best
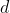 found. If a value larger than 0 is returned, it means that a
separable-plus-banded model can fit the data better compared to a
separable model. If we instead of fit-based CV decide to used
prediction-based CV, it makes sense to check the gains in prediction:
found. If a value larger than 0 is returned, it means that a
separable-plus-banded model can fit the data better compared to a
separable model. If we instead of fit-based CV decide to used
prediction-based CV, it makes sense to check the gains in prediction:
# X <- array(runif(100*20*30),c(100,20,30))
spb_est <- spb(X,predict=T)
spb$d
spb$cv
The same can be done with the
 -separable
model, replacing function
-separable
model, replacing function spb by scd, see ?spb and ?scd for a
more detailed description and examples. Note that by default, the mean
is always estimated empirically, unless other estimator of the mean is
provided.
Validity of a separable-plus-model can also be checked using a bootstrap
test, e.g. by
test_spb(X,d=1)
When one of the models is fitted, a list of functions that can be useful
to the user for subsequent manipulation of the estimated covariances is
as follows:
adi() solves the inverse problem involving a separable-plus-banded
covariancepcg() solves the inverse problem involving an
 -separable
covariance
-separable
covarianceapply_lhs() applies fast an
 -separable
covariance
-separable
covariance
To apply the separable-plus-banded model fast, see to_book_format and
BXfast. TODO:
- a handle for a fast application of the separable-plus-banded model
- a handle for best linear unbiased predictor (BLUP) using the
separable-plus-banded or
 -separable
model.
-separable
model.
TMasak/surfcov documentation built on April 25, 2022, 12:15 a.m.
surfcov
The purpose of R package surfcov is to enable covariance estimation
for random surfaces beyond separability, proposed in the papers
arXiv:1912.12870 and
arXiv:2007.12175.
Let
be i.i.d. matrices of size
representing discrete measurements (on a grid) of some latent random
surfaces on a 2D domain, and
be the covariance operator. The covariance is a tensor of size
,
which becomes problematic to handle for
and
as small as 100. The assumption of separability postulates that
which reduces statistical and computational burden, but is often critized as an oversimplification, since it does not allow any interaction between the two dimensions.
This package allows for efficient estimation and subsequent manipulation of two alternative models, which are both strict generalizations of separability:
- the separable-plus-banded model:
, where
for
or
;
- the
-separable model:
.
When the data are stored in form of an array X of size
,
it can be simply checked whether a given model can be potentially useful
by running
spb(X)for the separable-plus-banded model; orscd(X)for the-separable model.
Installation
You can install the development version from GitHub with:
install.packages("devtools") # only if devtools is not yet installed
library(devtools)
install_github("TMasak/surfcov")
Examples
When the data are stored in form of an array X of size
,
it can be simply checked whether a given model can be potentially useful
by running
spb(X)for the separable-plus-banded model; orscd(X)for the-separable model.
In particular,
# X <- array(runif(100*20*30),c(100,20,30))
spb_est <- spb(X)
spb$d
Run cross-validation (CV) in order to pick a value of the bandwidth
,
and then fits the model with the best
found. If a value larger than 0 is returned, it means that a
separable-plus-banded model can fit the data better compared to a
separable model. If we instead of fit-based CV decide to used
prediction-based CV, it makes sense to check the gains in prediction:
# X <- array(runif(100*20*30),c(100,20,30))
spb_est <- spb(X,predict=T)
spb$d
spb$cv
The same can be done with the
-separable
model, replacing function
spb by scd, see ?spb and ?scd for a
more detailed description and examples. Note that by default, the mean
is always estimated empirically, unless other estimator of the mean is
provided.
Validity of a separable-plus-model can also be checked using a bootstrap test, e.g. by
test_spb(X,d=1)
When one of the models is fitted, a list of functions that can be useful to the user for subsequent manipulation of the estimated covariances is as follows:
adi()solves the inverse problem involving a separable-plus-banded covariancepcg()solves the inverse problem involving an-separable covariance
apply_lhs()applies fast an-separable covariance
To apply the separable-plus-banded model fast, see to_book_format and
BXfast. TODO:
- a handle for a fast application of the separable-plus-banded model
- a handle for best linear unbiased predictor (BLUP) using the
separable-plus-banded or
-separable model.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
