README.md
In amalthomas111/SBF: R package to compute Shared Basis Factorization (SBF)
SBF: A R package for Shared Basis Factorization
Orthogonal Shared Basis Factorization (OSBF) is a joint matrix
diagonalization algorithm we developed for cross-species gene expression
analysis.
Installation
- If you have git installed, clone
SBF from Github and install.
git clone https://github.com/amalthomas111/SBF.git
If git is not installed, go to code -> Download ZIP. Unzip the file.
Then, inside an R console
install.packages("<path to SBF>/SBF", repos = NULL, type = "source")
[OR]
- Install using
devtools. Install devtools if not already
installed. Note: devtools requires lot of dependencies.
# check devtools is installed. If not install.
pkgs <- c("devtools")
require_install <- pkgs[!(pkgs %in% row.names(installed.packages()))]
if (length(require_install))
install.packages(require_install)
- Install SBF directly from Github via
devtools
library(devtools)
devtools::install_github("amalthomas111/SBF")
Please contact us via e-mail or submit a GitHub
issue if there is any
trouble with installation.
Quick demo
Load SBF package
# load SBF package
library(SBF)
Create a test dataset
- We will first create a test dataset.
SBF package has the function
createRandomMatrices to create matrices with full column rank and
different number of rows. All the matrices will have the same number
of columns.
set.seed(1231)
mymat <- createRandomMatrices(n = 4, ncols = 3, nrows = 4:6)
sapply(mymat, dim)
#> mat1 mat2 mat3 mat4
#> [1,] 5 6 4 5
#> [2,] 3 3 3 3
mymat
#> $mat1
#> [,1] [,2] [,3]
#> [1,] 47 98 10
#> [2,] 12 40 26
#> [3,] 60 62 41
#> [4,] 36 46 89
#> [5,] 53 61 83
#>
#> $mat2
#> [,1] [,2] [,3]
#> [1,] 12 92 42
#> [2,] 84 37 73
#> [3,] 38 33 29
#> [4,] 11 53 18
#> [5,] 74 41 6
#> [6,] 43 78 71
#>
#> $mat3
#> [,1] [,2] [,3]
#> [1,] 15 96 62
#> [2,] 87 81 59
#> [3,] 33 53 32
#> [4,] 3 69 7
#>
#> $mat4
#> [,1] [,2] [,3]
#> [1,] 3 39 97
#> [2,] 57 29 22
#> [3,] 20 89 58
#> [4,] 90 63 36
#> [5,] 35 71 13
Note: Depending upon the R versions, the random matrices generated could
be different.
The rank of the matrices
sapply(mymat, function(x) {qr(x)$rank})
#> mat1 mat2 mat3 mat4
#> 3 3 3 3
We will use the test dataset as our
 matrices.
matrices.
OSBF examples
The OSBF’s shared orthogonal
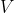 ,
,
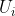 ’s
with orthonormal columns, and diagonal matrices
’s
with orthonormal columns, and diagonal matrices
 ’s
can be estimated using the
’s
can be estimated using the SBF function with argument
orthogonal = TRUE.
Check the arguments for the SBF function call using ?SBF.
# OSBF call
osbf <- SBF(matrix_list = mymat, orthogonal = TRUE)
#>
#> OSBF optimizing factorization error
names(osbf)
#> [1] "v" "u" "delta" "error"
#> [5] "error_pos" "error_vec" "v_start" "lambda_start"
#> [9] "u_start" "u_ortho_start" "delta_start" "m"
#> [13] "error_start"
For OSBF, the factorization error is optimized by default
(minimizeError=TRUE), and the optimizeFactorization function is
invoked. Optimization could take some time depending on the data matrix
(mymat) and initial values.
?SBF help function shows all arguments for the SBF call.
In OSBF,
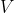 is orthogonal and columns of
is orthogonal and columns of
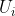 ’s
are orthonormal
(
’s
are orthonormal
( ).
).
zapsmall(t(osbf$v) %*% osbf$v)
#> [,1] [,2] [,3]
#> [1,] 1 0 0
#> [2,] 0 1 0
#> [3,] 0 0 1
# check the columns of first matrix of U_ortho
zapsmall(t(osbf$u[[names(osbf$u)[[1]]]]) %*%
osbf$u[[names(osbf$u)[[1]]]])
#> [,1] [,2] [,3]
#> [1,] 1 0 0
#> [2,] 0 1 0
#> [3,] 0 0 1
OSBF is not an exact factorization. The decomposition error is minimized
and the final factorization error is stored in osbf$error. The initial
factorization error we started with is stored in osbf$error_start.
# initial decomposition error
osbf$error_start
#> [1] 2329.73
# final decomposition error
osbf$error
#> [1] 1411.555
The error decreased by a factor of 1.65.
The number of iteration taken for optimizing:
cat("For osbf, # iteration =", osbf$error_pos, "final error =", osbf$error)
#> For osbf, # iteration = 220 final error = 1411.555
A detailed explanation of the algorithm and example cases of
factorization can be found in the vignettes/docs directory.
Factorization for an example gene expression dataset
- SBF package has a sample gene expression data with the mean
expression of nine tissues in five species. Now we will use this
data as our
 matrices.
matrices.
# load sample dataset from SBF package
avg_counts <- SBF::TissueExprSpecies
# dimension of matrices for different species
sapply(avg_counts, dim)
#> Homo_sapiens Macaca_mulatta Mus_musculus
#> [1,] 58676 30807 54446
#> [2,] 5 5 5
# names of different tissue types in humans
names(avg_counts[["Homo_sapiens"]])
#> [1] "hsapiens_brain" "hsapiens_heart" "hsapiens_kidney" "hsapiens_liver"
#> [5] "hsapiens_testis"
OSBF computation based on inter-sample correlation.
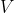 is estimated using the mean
is estimated using the mean
 .
.
# OSBF call using correlation matrix
osbf_gem <- SBF(matrix_list = avg_counts, orthogonal = TRUE,
transform_matrix = TRUE, tol = 1e-4)
#>
#> OSBF optimizing factorization error
# initial decomposition error
osbf_gem$error_start
#> [1] 65865.92
# final decomposition error
osbf_gem$error
#> [1] 7088.058
Note: For high-dimensional datasets, vary the tolerance threshold
(tol) or maximum number of iteration parameter (max_iter) to reduce
the computing time.
When calling SBF, orthogonal = FALSE computes the SBF factorization,
an exact joint matrix factorization we developed. In SBF, the estimated
columns of
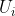 are not orthonormal. More details and examples can be found in the
vignettes/docs directory.
are not orthonormal. More details and examples can be found in the
vignettes/docs directory.
Contacts
Amal Thomas amalthom@usc.edu
Contact us via e-mail or submit a GitHub
issue
Publication
Available on
bioRxiv
Copyright
Copyright (C) 2018-2021 Amal Thomas
Authors: Amal Thomas
SBF is free software: you can redistribute it and/or modify it under the
terms of the GNU General Public License as published by the Free
Software Foundation, either version 3 of the License, or (at your
option) any later version.
SBF is distributed in the hope that it will be useful, but WITHOUT ANY
WARRANTY; without even the implied warranty of MERCHANTABILITY or
FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for
more details.
amalthomas111/SBF documentation built on Sept. 2, 2022, 11:27 a.m.
SBF: A R package for Shared Basis Factorization
Orthogonal Shared Basis Factorization (OSBF) is a joint matrix diagonalization algorithm we developed for cross-species gene expression analysis.
Installation
- If you have git installed, clone
SBFfrom Github and install.
git clone https://github.com/amalthomas111/SBF.git
If git is not installed, go to code -> Download ZIP. Unzip the file.
Then, inside an R console
install.packages("<path to SBF>/SBF", repos = NULL, type = "source")
[OR]
- Install using
devtools. Installdevtoolsif not already installed. Note:devtoolsrequires lot of dependencies.
# check devtools is installed. If not install.
pkgs <- c("devtools")
require_install <- pkgs[!(pkgs %in% row.names(installed.packages()))]
if (length(require_install))
install.packages(require_install)
- Install SBF directly from Github via
devtools
library(devtools)
devtools::install_github("amalthomas111/SBF")
Please contact us via e-mail or submit a GitHub issue if there is any trouble with installation.
Quick demo
Load SBF package
# load SBF package
library(SBF)
Create a test dataset
- We will first create a test dataset.
SBFpackage has the functioncreateRandomMatricesto create matrices with full column rank and different number of rows. All the matrices will have the same number of columns.
set.seed(1231)
mymat <- createRandomMatrices(n = 4, ncols = 3, nrows = 4:6)
sapply(mymat, dim)
#> mat1 mat2 mat3 mat4
#> [1,] 5 6 4 5
#> [2,] 3 3 3 3
mymat
#> $mat1
#> [,1] [,2] [,3]
#> [1,] 47 98 10
#> [2,] 12 40 26
#> [3,] 60 62 41
#> [4,] 36 46 89
#> [5,] 53 61 83
#>
#> $mat2
#> [,1] [,2] [,3]
#> [1,] 12 92 42
#> [2,] 84 37 73
#> [3,] 38 33 29
#> [4,] 11 53 18
#> [5,] 74 41 6
#> [6,] 43 78 71
#>
#> $mat3
#> [,1] [,2] [,3]
#> [1,] 15 96 62
#> [2,] 87 81 59
#> [3,] 33 53 32
#> [4,] 3 69 7
#>
#> $mat4
#> [,1] [,2] [,3]
#> [1,] 3 39 97
#> [2,] 57 29 22
#> [3,] 20 89 58
#> [4,] 90 63 36
#> [5,] 35 71 13
Note: Depending upon the R versions, the random matrices generated could be different.
The rank of the matrices
sapply(mymat, function(x) {qr(x)$rank})
#> mat1 mat2 mat3 mat4
#> 3 3 3 3
We will use the test dataset as our
matrices.
OSBF examples
The OSBF’s shared orthogonal
,
’s
with orthonormal columns, and diagonal matrices
’s
can be estimated using the
SBF function with argument
orthogonal = TRUE.
Check the arguments for the SBF function call using ?SBF.
# OSBF call
osbf <- SBF(matrix_list = mymat, orthogonal = TRUE)
#>
#> OSBF optimizing factorization error
names(osbf)
#> [1] "v" "u" "delta" "error"
#> [5] "error_pos" "error_vec" "v_start" "lambda_start"
#> [9] "u_start" "u_ortho_start" "delta_start" "m"
#> [13] "error_start"
For OSBF, the factorization error is optimized by default
(minimizeError=TRUE), and the optimizeFactorization function is
invoked. Optimization could take some time depending on the data matrix
(mymat) and initial values.
?SBF help function shows all arguments for the SBF call.
In OSBF,
is orthogonal and columns of
’s
are orthonormal
(
).
zapsmall(t(osbf$v) %*% osbf$v)
#> [,1] [,2] [,3]
#> [1,] 1 0 0
#> [2,] 0 1 0
#> [3,] 0 0 1
# check the columns of first matrix of U_ortho
zapsmall(t(osbf$u[[names(osbf$u)[[1]]]]) %*%
osbf$u[[names(osbf$u)[[1]]]])
#> [,1] [,2] [,3]
#> [1,] 1 0 0
#> [2,] 0 1 0
#> [3,] 0 0 1
OSBF is not an exact factorization. The decomposition error is minimized
and the final factorization error is stored in osbf$error. The initial
factorization error we started with is stored in osbf$error_start.
# initial decomposition error
osbf$error_start
#> [1] 2329.73
# final decomposition error
osbf$error
#> [1] 1411.555
The error decreased by a factor of 1.65.
The number of iteration taken for optimizing:
cat("For osbf, # iteration =", osbf$error_pos, "final error =", osbf$error)
#> For osbf, # iteration = 220 final error = 1411.555
A detailed explanation of the algorithm and example cases of factorization can be found in the vignettes/docs directory.
Factorization for an example gene expression dataset
- SBF package has a sample gene expression data with the mean
expression of nine tissues in five species. Now we will use this
data as our
matrices.
# load sample dataset from SBF package
avg_counts <- SBF::TissueExprSpecies
# dimension of matrices for different species
sapply(avg_counts, dim)
#> Homo_sapiens Macaca_mulatta Mus_musculus
#> [1,] 58676 30807 54446
#> [2,] 5 5 5
# names of different tissue types in humans
names(avg_counts[["Homo_sapiens"]])
#> [1] "hsapiens_brain" "hsapiens_heart" "hsapiens_kidney" "hsapiens_liver"
#> [5] "hsapiens_testis"
OSBF computation based on inter-sample correlation.
is estimated using the mean
.
# OSBF call using correlation matrix
osbf_gem <- SBF(matrix_list = avg_counts, orthogonal = TRUE,
transform_matrix = TRUE, tol = 1e-4)
#>
#> OSBF optimizing factorization error
# initial decomposition error
osbf_gem$error_start
#> [1] 65865.92
# final decomposition error
osbf_gem$error
#> [1] 7088.058
Note: For high-dimensional datasets, vary the tolerance threshold
(tol) or maximum number of iteration parameter (max_iter) to reduce
the computing time.
When calling SBF, orthogonal = FALSE computes the SBF factorization,
an exact joint matrix factorization we developed. In SBF, the estimated
columns of
are not orthonormal. More details and examples can be found in the
vignettes/docs directory.
Contacts
Amal Thomas amalthom@usc.edu
Contact us via e-mail or submit a GitHub issue
Publication
Available on bioRxiv
Copyright
Copyright (C) 2018-2021 Amal Thomas
Authors: Amal Thomas
SBF is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
SBF is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
