In ardata-fr/dgihesse: Simple 'JavaScript' Interactions in 'shiny' Applications
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
fig.path = "man/figures/README-",
out.width = "100%"
)
shinytools
 shinytools brings some minor but important features in shiny applications by providing
simple JavaScript functions to make interactions with the DOM easier and modules
to perform data importation and data filtering in shiny applications.
shinytools brings some minor but important features in shiny applications by providing
simple JavaScript functions to make interactions with the DOM easier and modules
to perform data importation and data filtering in shiny applications.
The first motivation of shinytools is to gather and share codes written by ArData
when building Shiny applications.
JavaScript functions
The package is providing JavaScript bindings for common and useful operations as shiny utilities :
- disable or enable a shiny control:
ability(), html_disable(), html_enable(), default_disabled()
- display or hide an HTML element:
html_toogle(), html_set_visible(), html_set_hidden()
- set or unset active state for a button:
activate(), html_set_active(), html_set_inactive()
- create a reactive value from a click event:
click_event
- add or remove a class:
html_class(), html_addclass(), html_unclass()
Simple shiny modules
The package also provides some of the modules we use :
- A tool for data importation:
importDataUI & importDataServer
- A tool for data filtering:
filterDataUI & filterDataServer
Installation
# install.packages("remotes")
remotes::install_github("ardata-fr/shinytools")
Example
Disable inputs
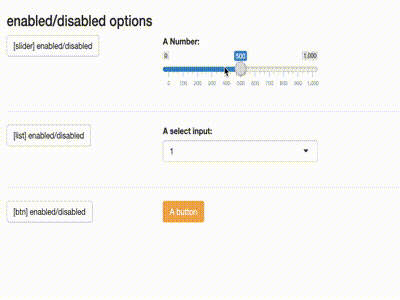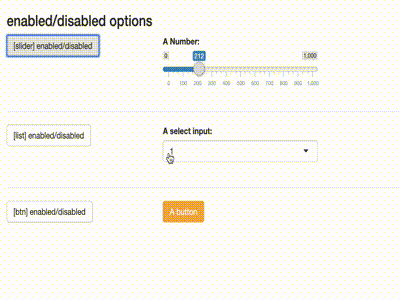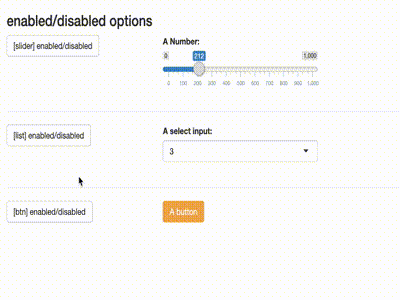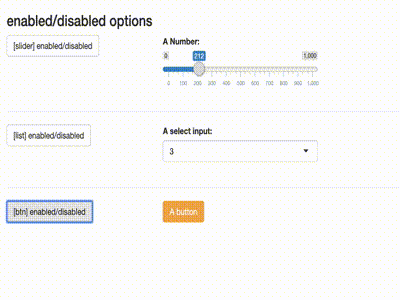
library(shiny)
library(shinytools)
if (interactive()) {
ui <- fluidPage(
load_jstools(),
fluidRow(column(width = 12, h3("enabled/disabled options"))),
fluidRow(
column(width = 3,
actionButton(inputId = "able_slider",
label = "[slider] enabled/disabled") ),
column(width = 5,
sliderInput( "slider",
"A Number:",
min = 0, max = 1000, value = 500)
)
),
hr(),
fluidRow(
column(width = 3,
actionButton(inputId = "able_select",
label = "[list] enabled/disabled")),
column(width = 5,
selectizeInput("select", "A select input:", 1:5)
)
),
hr(),
fluidRow(
column(width = 3,
actionButton(inputId = "able_btn",
label = "[btn] enabled/disabled")),
column(width = 5,
actionButton("btn", "A button", class = "btn-warning")
)
)
)
server <- function(input, output) {
observeEvent(input$able_slider, {
ability("slider", input$able_slider%%2 < 1)
})
observeEvent(input$able_btn, {
ability("btn", input$able_btn%%2 < 1)
})
observeEvent(input$able_select, {
ability("select", input$able_select%%2 < 1)
})
}
print(shinyApp(ui, server))
}
Import data

if (interactive()) {
options(device.ask.default = FALSE)
ui <- fluidPage(
titlePanel("Import and visualize dataset"),
sidebarLayout(
sidebarPanel(
load_tingle(),
importDataUI(id = "id1"),
uiOutput("dataset_labels")
),
mainPanel(
DT::dataTableOutput(outputId = "id2")
)
)
)
server <- function(input, output) {
all_datasets <- reactiveValues()
datasets <- callModule(
module = importDataServer,
id = "id1", ui_element = "actionButton",
labelize = TRUE,
forbidden_labels = reactive(names(reactiveValuesToList(all_datasets))))
observeEvent(datasets$trigger, {
req(datasets$trigger > 0)
all_datasets[[datasets$name]] <- datasets$object
})
output$dataset_labels <- renderUI({
x <- reactiveValuesToList(all_datasets)
if (length(x) > 0) {
selectInput("SI_labels", label = "Choose dataset", choices = names(x))
}
})
output$id2 <- DT::renderDataTable({
req(input$SI_labels)
all_datasets[[input$SI_labels]]
})
}
print(shinyApp(ui, server))
}
Filter data
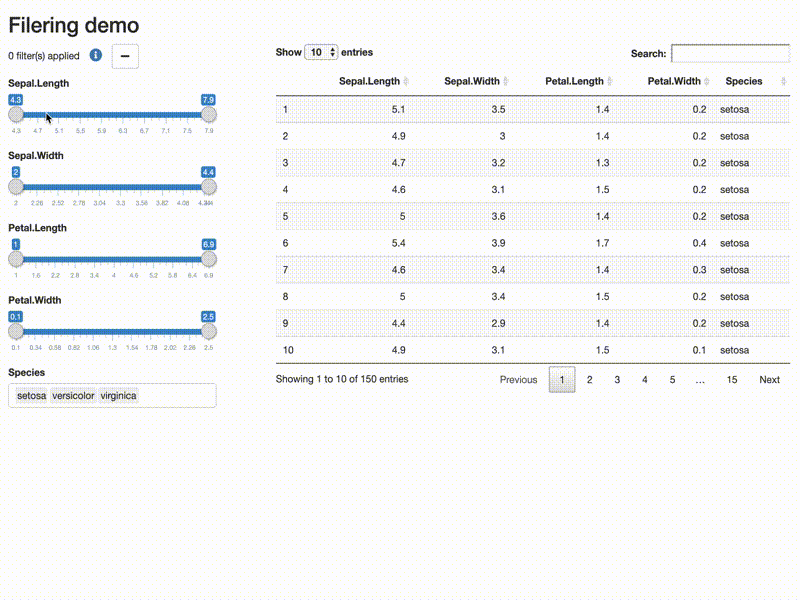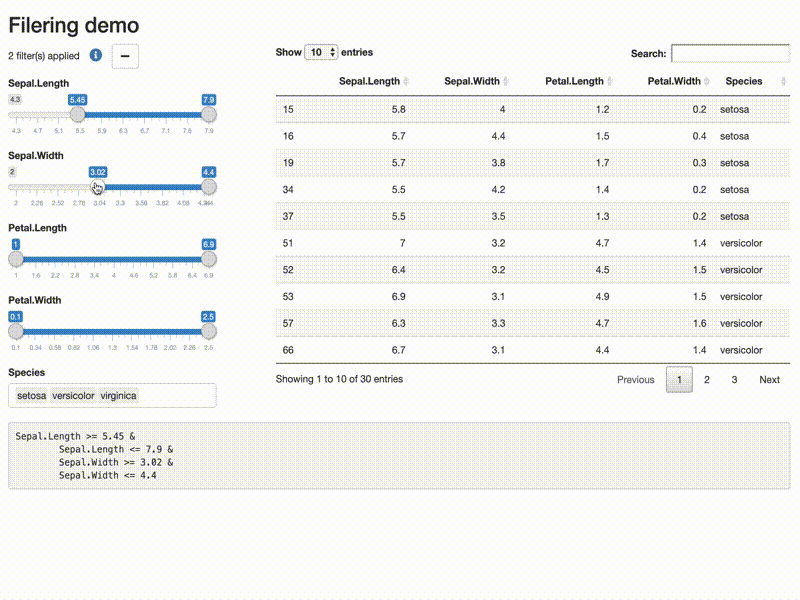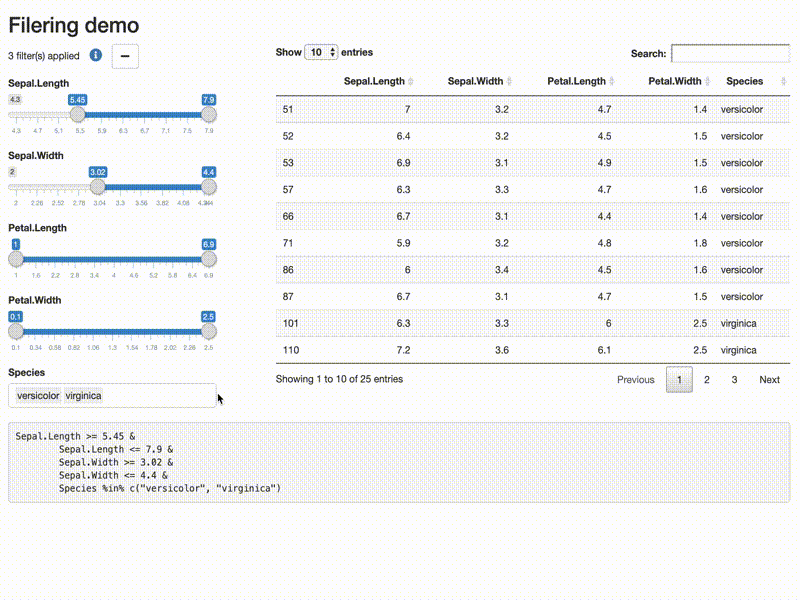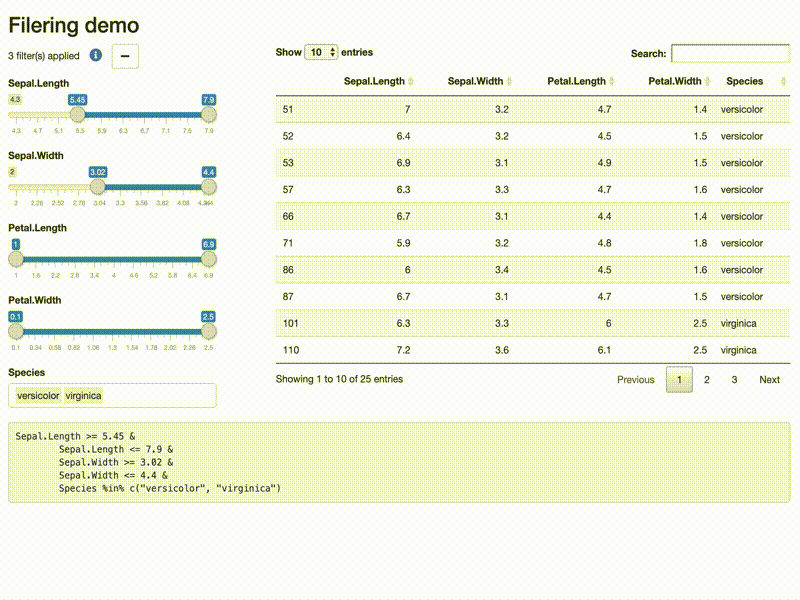
library(shiny)
library(DT)
library(shinytools)
if (interactive()) {
options(device.ask.default = FALSE)
ui <- fluidPage(
fluidRow(column(width=12, h2("Filering demo"))),
fluidRow(
column(
width = 4,
filterDataUI(id = "demo")
),
column(width = 8,
DT::dataTableOutput(outputId = "subsetdata")
)
),
fluidRow(
column(width = 12,
verbatimTextOutput(outputId = "expr")
)
)
)
server <- function(input, output, session) {
res <- callModule(module = filterDataServer,
id = "demo", x = reactive(iris),
return_data = TRUE)
output$expr <- renderText({
req(res)
if(res$filtered){
expr_str <- format(res$expr)
expr_str <- paste( gsub("^[ ]+", "", expr_str), collapse = "")
gsub("\\&[ ]*", "&\n\t", expr_str, fixed = FALSE)
} else NULL
})
output$subsetdata <- DT::renderDataTable({
res$filtered_data
})
}
print(shinyApp(ui, server))
}
If you set the parameter return_data = FALSE then you can evaluate the returned call as follow :
# With base R
filters <- eval(expr = res$expr, envir = iris)
# With lazyeval
filters <- lazyeval::lazy_eval(res$expr, data = iris)
# With rlang
filters <- rlang::eval_tidy(res$expr, data = iris)
# Then subset data.frame
iris[filters,]
ardata-fr/dgihesse documentation built on Nov. 14, 2019, 7:25 a.m.
knitr::opts_chunk$set( collapse = TRUE, comment = "#>", fig.path = "man/figures/README-", out.width = "100%" )
shinytools

The first motivation of shinytools is to gather and share codes written by ArData when building Shiny applications.
JavaScript functions
The package is providing JavaScript bindings for common and useful operations as shiny utilities :
- disable or enable a shiny control:
ability(),html_disable(),html_enable(),default_disabled() - display or hide an HTML element:
html_toogle(),html_set_visible(),html_set_hidden() - set or unset active state for a button:
activate(),html_set_active(),html_set_inactive() - create a reactive value from a click event:
click_event - add or remove a class:
html_class(),html_addclass(),html_unclass()
Simple shiny modules
The package also provides some of the modules we use :
- A tool for data importation:
importDataUI&importDataServer - A tool for data filtering:
filterDataUI&filterDataServer
Installation
# install.packages("remotes") remotes::install_github("ardata-fr/shinytools")
Example
Disable inputs

library(shiny) library(shinytools) if (interactive()) { ui <- fluidPage( load_jstools(), fluidRow(column(width = 12, h3("enabled/disabled options"))), fluidRow( column(width = 3, actionButton(inputId = "able_slider", label = "[slider] enabled/disabled") ), column(width = 5, sliderInput( "slider", "A Number:", min = 0, max = 1000, value = 500) ) ), hr(), fluidRow( column(width = 3, actionButton(inputId = "able_select", label = "[list] enabled/disabled")), column(width = 5, selectizeInput("select", "A select input:", 1:5) ) ), hr(), fluidRow( column(width = 3, actionButton(inputId = "able_btn", label = "[btn] enabled/disabled")), column(width = 5, actionButton("btn", "A button", class = "btn-warning") ) ) ) server <- function(input, output) { observeEvent(input$able_slider, { ability("slider", input$able_slider%%2 < 1) }) observeEvent(input$able_btn, { ability("btn", input$able_btn%%2 < 1) }) observeEvent(input$able_select, { ability("select", input$able_select%%2 < 1) }) } print(shinyApp(ui, server)) }
Import data

if (interactive()) { options(device.ask.default = FALSE) ui <- fluidPage( titlePanel("Import and visualize dataset"), sidebarLayout( sidebarPanel( load_tingle(), importDataUI(id = "id1"), uiOutput("dataset_labels") ), mainPanel( DT::dataTableOutput(outputId = "id2") ) ) ) server <- function(input, output) { all_datasets <- reactiveValues() datasets <- callModule( module = importDataServer, id = "id1", ui_element = "actionButton", labelize = TRUE, forbidden_labels = reactive(names(reactiveValuesToList(all_datasets)))) observeEvent(datasets$trigger, { req(datasets$trigger > 0) all_datasets[[datasets$name]] <- datasets$object }) output$dataset_labels <- renderUI({ x <- reactiveValuesToList(all_datasets) if (length(x) > 0) { selectInput("SI_labels", label = "Choose dataset", choices = names(x)) } }) output$id2 <- DT::renderDataTable({ req(input$SI_labels) all_datasets[[input$SI_labels]] }) } print(shinyApp(ui, server)) }
Filter data

library(shiny) library(DT) library(shinytools) if (interactive()) { options(device.ask.default = FALSE) ui <- fluidPage( fluidRow(column(width=12, h2("Filering demo"))), fluidRow( column( width = 4, filterDataUI(id = "demo") ), column(width = 8, DT::dataTableOutput(outputId = "subsetdata") ) ), fluidRow( column(width = 12, verbatimTextOutput(outputId = "expr") ) ) ) server <- function(input, output, session) { res <- callModule(module = filterDataServer, id = "demo", x = reactive(iris), return_data = TRUE) output$expr <- renderText({ req(res) if(res$filtered){ expr_str <- format(res$expr) expr_str <- paste( gsub("^[ ]+", "", expr_str), collapse = "") gsub("\\&[ ]*", "&\n\t", expr_str, fixed = FALSE) } else NULL }) output$subsetdata <- DT::renderDataTable({ res$filtered_data }) } print(shinyApp(ui, server)) }
If you set the parameter return_data = FALSE then you can evaluate the returned call as follow :
# With base R filters <- eval(expr = res$expr, envir = iris) # With lazyeval filters <- lazyeval::lazy_eval(res$expr, data = iris) # With rlang filters <- rlang::eval_tidy(res$expr, data = iris) # Then subset data.frame iris[filters,]
Embedding an R snippet on your website
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
