README.md
In aseyq/flyCSV: Opens your data frame as CSV
flyCSV: Check your data frame as CSV on the fly! (R Package)
Warning: This is a very preliminary version.
This library (well, more a function) helps you view your data frames CSV files on the fly. It is basically a wrapper that saves the database temporarily, and then opens it with the default program. The functionality is similar to the built-in View() function, however, unlike View() it returns the same object, so it can be used in between pipe chains. Also it doesn't rely on RStudio.
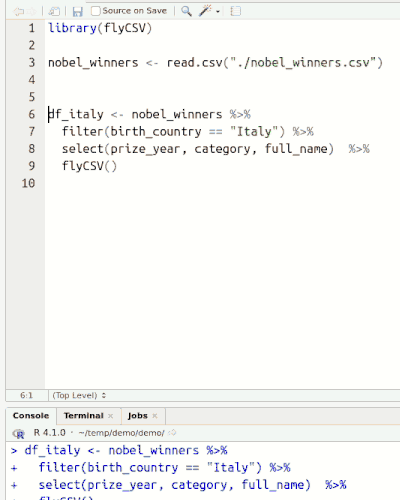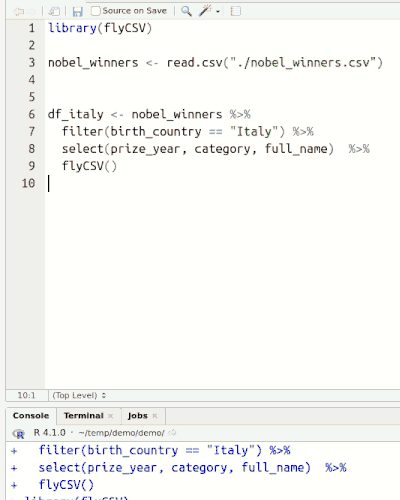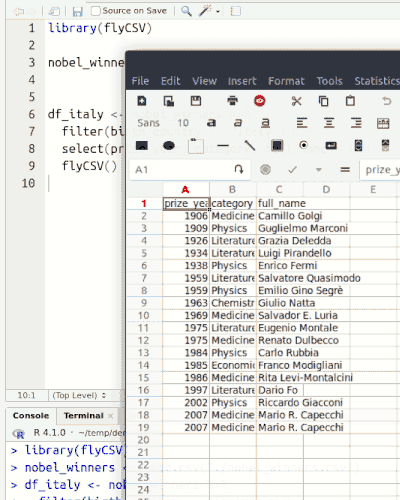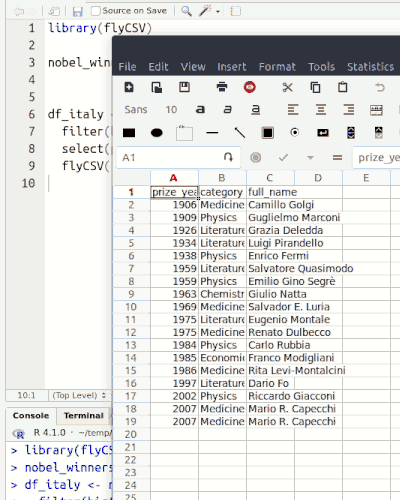
Installation
You can install it by:
devtools::install_github("aseyq/flyCSV")
Devtools package is necessary if you want to install an R package directly from github. You probably already have it but if you don't, you can install it with:
install.packages("devtools")
Usage
Load the library
library(flycsv)
Basic Usage
flyCSV(df)
You can use it at the end of a pipe
magrittr pipes
df %>%
somefunction(...) %>%
flyCSV()
Base R pipes (from R 4.1 on)
df |>
somefunction(...) |>
flyCSV()
as well as between pipes
df %>%
somefunction(...) %>%
flyCSV() %>% # My csv will show the changes up to this point!
someotherfunction(...)
you can save your data while using flyCSV
new_df <- df %>%
somefunction(...) %>%
flyCSV()
Write the file with a specific name in a directory
In this case it won't be deleted automatically.
df %>%
somefunction(...) %>%
flyCSV("my_file.csv")
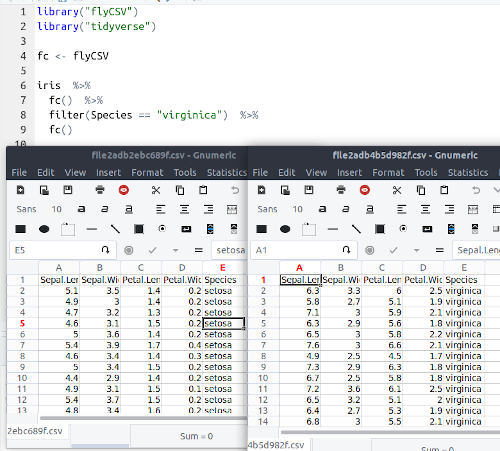
It supports calling multiple times, so it can be useful to compare the data
df %>%
do_something() %>%
flyCSV("before.csv") %>%
do_something_else() %>%
flyCSV("after.csv")
Change the software to open the file
df %>%
somefunction(...) %>%
flyCSV("my_file.csv", browser="C:\Program Files\LibreOffice\program\soffice.exe")
Tip: Using flyDN (stands for Do Nothing) to quick comment-outs and ins
Let's say that you are working on a long pipe and time to time you want to investigate your
data at the end of a pipe by commenting out and in flyCSV function. With pipes, if you
comment the pipe at the end of the chain, since the previous pipe will not have function to
input, you will get an error, or your R will wait for an input. For instance:
- This will not work unless you remove the pipe in the previous line:
iris %>%
filter(Species == "virginica") %>%
# flyCSV()
To facilitate commenting ins and outs, flyCSV comes with a function flyDN which does nothing but
returns the same dataframe. Therefore you can put flyDN() at the end of your pipes to uncomment
and comment easily the function before. For insance:
- This will work:
iris %>%
filter(Species == "virginica") %>%
# flyCSV() %>%
flyDN()
- As well as this one:
iris %>%
filter(Species == "virginica") %>%
flyCSV() %>%
flyDN()
Tip: Using an alias
You can create an alias for flyCSV to speed your writing up when you are investigating your data.
fc <- flyCSV
df %>%
do_something() %>%
fc()
For example
Input types and output structures
flyCSV uses write.csv underneath, which is extremely flexible. So,flyCSV() can take different types of input. Here is a demonstration of how the CSV file structure would look like for different formats.
Input is a vector
my_vector <- c(1,2,3,4)
flyCSV(my_vector)
| x |
| - |
| 1 |
| 2 |
| 3 |
| 4 |
Input is a data.frame (or tibble, or data.table)
my_df <- data.frame(a=c(1,2,3,4), b=c(4,3,2,1))
flyCSV(my_df)
| a | b |
| - | - |
| 1 | 4 |
| 2 | 3 |
| 3 | 2 |
| 4 | 1 |
Input is a matrix
my_matrix <- matrix(c(1,2,3,4,5,6), nrow=2)
flyCSV(my_matrix)
| V1 | V2 | V3 |
| -- | -- | -- |
| 1 | 3 | 5 |
| 2 | 4 | 6 |
Input is a list of data.frames (or tibbles, or data.tables)
df1 <- data.frame(a=c(1,2,3,4), b=c(4,3,2,1))
df2 <- data.frame(a=c(1,3,2,4), b=c(4,2,1,3))
my_list_of_dfs <- list(df1=df1, df2=df2)
flyCSV(my_list_of_dfs)
| df1.a | df1.b | df2.a | df2.b |
| ----- | ----- | ----- | ----- |
| 1 | 4 | 1 | 4 |
| 2 | 3 | 3 | 2 |
| 3 | 2 | 2 | 1 |
| 4 | 1 | 4 | 3 |
aseyq/flyCSV documentation built on Aug. 6, 2022, 10:06 p.m.
flyCSV: Check your data frame as CSV on the fly! (R Package)
Warning: This is a very preliminary version.

This library (well, more a function) helps you view your data frames CSV files on the fly. It is basically a wrapper that saves the database temporarily, and then opens it with the default program. The functionality is similar to the built-in View() function, however, unlike View() it returns the same object, so it can be used in between pipe chains. Also it doesn't rely on RStudio.

Installation
You can install it by:
devtools::install_github("aseyq/flyCSV")
Devtools package is necessary if you want to install an R package directly from github. You probably already have it but if you don't, you can install it with:
install.packages("devtools")
Usage
Load the library
library(flycsv)
Basic Usage
flyCSV(df)
You can use it at the end of a pipe
magrittr pipes
df %>%
somefunction(...) %>%
flyCSV()
Base R pipes (from R 4.1 on)
df |>
somefunction(...) |>
flyCSV()
as well as between pipes
df %>%
somefunction(...) %>%
flyCSV() %>% # My csv will show the changes up to this point!
someotherfunction(...)
you can save your data while using flyCSV
new_df <- df %>%
somefunction(...) %>%
flyCSV()
Write the file with a specific name in a directory
In this case it won't be deleted automatically.
df %>%
somefunction(...) %>%
flyCSV("my_file.csv")
It supports calling multiple times, so it can be useful to compare the data
df %>%
do_something() %>%
flyCSV("before.csv") %>%
do_something_else() %>%
flyCSV("after.csv")
Change the software to open the file
df %>%
somefunction(...) %>%
flyCSV("my_file.csv", browser="C:\Program Files\LibreOffice\program\soffice.exe")
Tip: Using flyDN (stands for Do Nothing) to quick comment-outs and ins
Let's say that you are working on a long pipe and time to time you want to investigate your
data at the end of a pipe by commenting out and in flyCSV function. With pipes, if you
comment the pipe at the end of the chain, since the previous pipe will not have function to
input, you will get an error, or your R will wait for an input. For instance:
- This will not work unless you remove the pipe in the previous line:
iris %>%
filter(Species == "virginica") %>%
# flyCSV()
To facilitate commenting ins and outs, flyCSV comes with a function flyDN which does nothing but
returns the same dataframe. Therefore you can put flyDN() at the end of your pipes to uncomment
and comment easily the function before. For insance:
- This will work:
iris %>%
filter(Species == "virginica") %>%
# flyCSV() %>%
flyDN()
- As well as this one:
iris %>%
filter(Species == "virginica") %>%
flyCSV() %>%
flyDN()
Tip: Using an alias
You can create an alias for flyCSV to speed your writing up when you are investigating your data.
fc <- flyCSV
df %>%
do_something() %>%
fc()
For example

Input types and output structures
flyCSV uses write.csv underneath, which is extremely flexible. So,flyCSV() can take different types of input. Here is a demonstration of how the CSV file structure would look like for different formats.
Input is a vector
my_vector <- c(1,2,3,4)
flyCSV(my_vector)
| x | | - | | 1 | | 2 | | 3 | | 4 |
Input is a data.frame (or tibble, or data.table)
my_df <- data.frame(a=c(1,2,3,4), b=c(4,3,2,1))
flyCSV(my_df)
| a | b | | - | - | | 1 | 4 | | 2 | 3 | | 3 | 2 | | 4 | 1 |
Input is a matrix
my_matrix <- matrix(c(1,2,3,4,5,6), nrow=2)
flyCSV(my_matrix)
| V1 | V2 | V3 | | -- | -- | -- | | 1 | 3 | 5 | | 2 | 4 | 6 |
Input is a list of data.frames (or tibbles, or data.tables)
df1 <- data.frame(a=c(1,2,3,4), b=c(4,3,2,1))
df2 <- data.frame(a=c(1,3,2,4), b=c(4,2,1,3))
my_list_of_dfs <- list(df1=df1, df2=df2)
flyCSV(my_list_of_dfs)
| df1.a | df1.b | df2.a | df2.b | | ----- | ----- | ----- | ----- | | 1 | 4 | 1 | 4 | | 2 | 3 | 3 | 2 | | 3 | 2 | 2 | 1 | | 4 | 1 | 4 | 3 |
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
