README.md
In luluperet/icor: What the Package Does (One Line, Title Case)
ICOR, Correlation and Usefull Functions, Operators and Classes
EXAMPLES
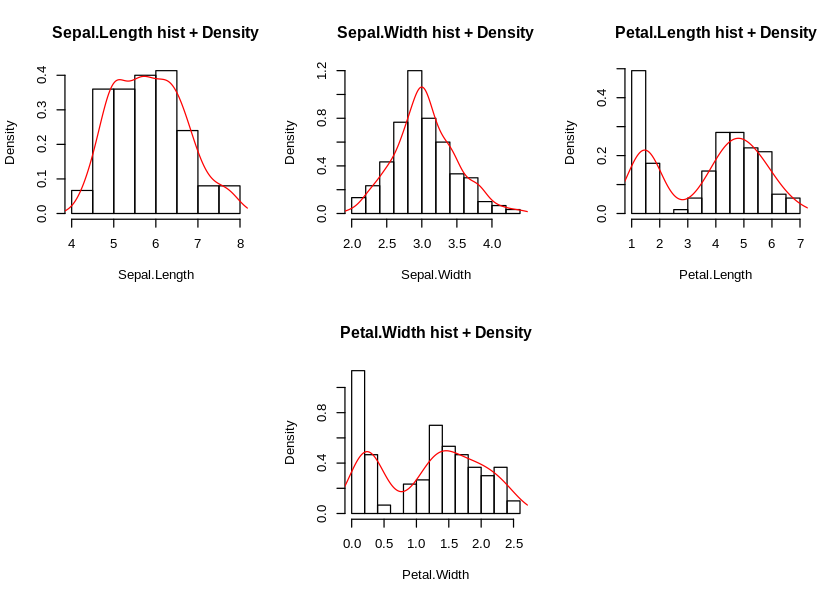
iris %>%
numericCol %getCol%
1:4 %>skip>% {
layout_build_matrix(
1,2,3|
0,4,0
);
resetPlt = plotWH(7,5)
}%each% l__(
{
name=names(dimnames(.))
hist(.x,xlab = name,main = name%.%" hist + Density",probability = T);
densityLines(.x,col="red")
}
) %>% invisible
#SAME
#We use eachCol instead of each (.y is the colname)
#We use tg instead of invisible (same, but remove warnings too)
iris %>%
numericCol %getCol%
1:4 %>skip>% {
layout_build_matrix(
1,2,3|
0,4,0
);
resetPlt = plotWH(7,5)
} %eachCol% l1__(
{
name=.y
hist(.x,xlab = name,main = name%.%" hist + Density",probability = T);
densityLines(.x,col="red")
}
) %>% tg
EQ
Test values each in vector
< <= == >= >
%||%
```R
c(4,2,1) %||% c(9,4,0) < 3
[1] FALSE
**%|%**</br>R
c(4,2,1) %|% c(9,4,0) < 3
[1] FALSE TRUE TRUE
**%&&%** </br>R
c(4,2,1) %&&% c(9,4,0) < 3
[1] FALSE
**%&%**</br>R
c(4,2,1) %&% c(9,4,0) < 3
[1] FALSE FALSE TRUE
```
list To Parameters of a Function
Call function of the right side, with elements of the list passed in the left side
Operators
%...>%(#list|#atomic,#function) (keep names)
```R
list(mean=10,n=5,sd = 2) %...>% rnorm
[1] 12.817348 10.284119 11.355081 7.372176 7.795275
list(mean=10,n=5,sd = 2) %...>% smth
List of 3
$ mean: num 10
$ n : num 5
$ sd : num 2
10 %,% 20 %...>% rnorm
[1] 19.51830 19.32193 17.89852 20.27248 19.79469 22.44875 21.06525 22.81541 20.42578
[10] 20.35683
(see #icor_lists below)
10 %...>% rnorm
[1] 0.88250520 -1.06436486 0.44685920 -0.58315920 1.05593594 0.28902886 -0.01550811
[8] 0.76758399 2.07238604 -0.86256077
**%..._>%(#list|#atomic,#function) (not keep names)**R
list(mean=10,n=5,sd = 2) %..._>% rnorm
[1] 4.5024198 8.4893204 10.0372266 4.5468650 3.7732437 4.1219222
[7] 3.7688349 0.1318678 0.9097222 7.8097455
list(mean=10,n=5,sd = 2) %..._>% smth
List of 3
$ : num 10
$ : num 5
$ : num 2
10 %,% 20 %...>% rnorm
[1] 20.33436 19.57912 19.15201 18.57136 19.53694 19.09004 18.62865 19.14920 19.08933
[10] 19.00091
#(see #icor_lists below)
10 %..._>% rnorm
[1] 0.88250520 -1.06436486 0.44685920 -0.58315920 1.05593594 0.28902886 -0.01550811
[8] 0.76758399 2.07238604 -0.86256077
**%listToDotsFn%(#list,#function) (keep names)**R
list(mean=10,n=5,sd = 2) %listToDotsFn% rnorm
[1] 11.396479 10.744122 8.012919 11.297808 12.601390
list(ind="a",day=3,month=4) %listToDotsFn% smth
List of 3
$ ind : chr "a"
$ day : num 3
$ month: num 4
**%listToDotsFn_%(#list,#function) (not keep names)**
*(see "smth" below)*</br>R
list(ind="a",day=3,month=4) %listToDotsFn_% smth
List of 3
$ : chr "a"
$ : num 3
$ : num 4
10 %,% 20 %listToDotsFn_% rnorm
[1] 20.33436 19.57912 19.15201 18.57136 19.53694 19.09004 18.62865 19.14920 19.08933
[10] 19.00091
(see #icor_lists below)
```
String Concat
%.=%
Concat and assign Strings
> a="Hello "
> a %.=% "World"
> cat(a)
Hello World
%.%
Concat strings (paste0)
```R
"ll"%.%"kk"%.%"kkd"
[1] "llkkkkd"
```
Usefull Utils
curry(fn)
(Currying)[https://en.wikipedia.org/wiki/Currying]
```R
a=curry(rnorm(10))
a
function (...)
rnorm(10, ...)
a(10)
[1] 9.822035 9.897887 10.289826 9.588149 9.705572 11.554598
[7] 11.485561 8.851399 12.550492 10.740860
**smth**</br>
*Return str of list of dots* w/br>R
3 %,% 4 %,% l(d=3) %->% smth
List of 3
$ : num 3
$ : num 4
$ :List of 1
..$ d: num 3
smth(1,2,3,4,5,"kkk",rnorm,l1___(rnorm(10)))
List of 8
$ : num 1
$ : num 2
$ : num 3
$ : num 4
$ : num 5
$ : chr "kkk"
$ :function (n, mean = 0, sd = 1)
$ :function (...)
..- attr(, "class")= chr [1:2] "purrr_function_partial" "function"
..- attr(, "body")= language ~(function (n, mean = 0, sd = 1) .Call(C_rnorm, n, mean, sd))(10, ...)
.. ..- attr(, ".Environment")=
..- attr(, "fn")= symbol rnorm
**startsWithFromList(list,string)**</br>
*filter list with element who begin with string*</br>R
c("hello","he lo","hehe","bonjour","au revoir") %>% startsWithFromList("he")
[1] "hello" "he lo" "hehe"
**addNamesToList(list,names)**</br>
*add names to list*</br>R
c("hello","he lo","hehe","bonjour","au revoir") %>% addNamesToList(1:5)
1 2 3 4 5
"hello" "he lo" "hehe" "bonjour" "au revoir"
c("hello","he lo","hehe","bonjour","au revoir") %>% addNamesToList(1:5 %each% {"d"%.%.})
d1 d2 d3 d4 d5
"hello" "he lo" "hehe" "bonjour" "au revoir"
l("hello","he lo","hehe","bonjour","au revoir") %>% addNamesToList(1:5 %each% {"d"%.%.})
$d1
[1] "hello"
$d2
[1] "he lo"
$d3
[1] "hehe"
$d4
[1] "bonjour"
$d5
[1] "au revoir"
```
nothing(...)
return nothing with whatever params
%call%(#fn,#list)
*call fn with elements of list as parameters
%>skip>%(data,#fn) doAndSkip %-|skip|->%
Execute fn and return data ! all above do the same
Usefull DataFrame
toDF(list) toDFt(list)
convert an list to a dataframe with good colnames, rownames
```R
l("hello","he lo","hehe","bonjour","au revoir") %>% addNamesToList(1:5 %each% {"d"%.%.}) %>% toDF
[,1]
d1 "hello"
d2 "he lo"
d3 "hehe"
d4 "bonjour"
d5 "au revoir"
l("hello","he lo","hehe","bonjour","au revoir") %>% addNamesToList(1:5 %each% {"d"%.%.}) %>% toDFt
d1 d2 d3 d4 d5
1 hello he lo hehe bonjour au revoir
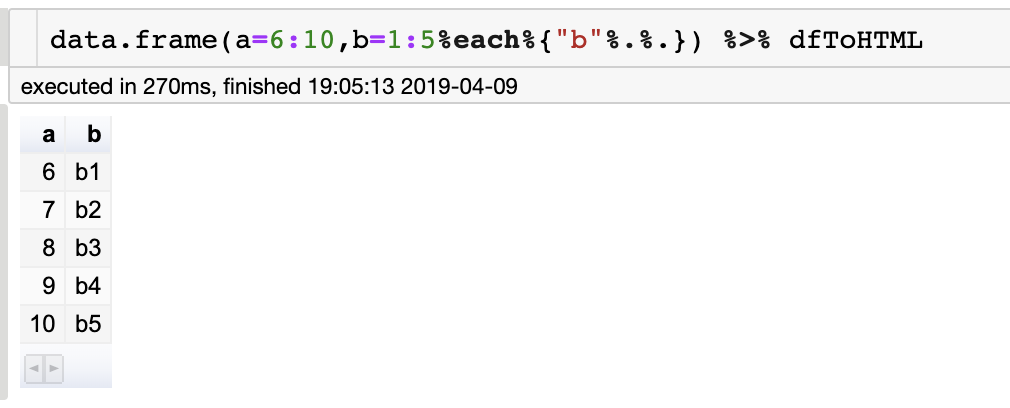
**add_row_with_name(#data.frame)**</br>
*add row with rowname in a data frame*</br>R
data.frame(a=6:10,b=1:5%each%{"b"%.%.})
a b
1 6 b1
2 7 b2
3 8 b3
4 9 b4
5 10 b5
data.frame(a=6:10,b=1:5%each%{"b"%.%.}) %>% add_row_with_name(a=9,b="b5",name="9")
a b
1 6 b1
2 7 b2
3 8 b3
4 9 b4
5 10 b5
9 9 b5
**dfRowToList**</br>
*convert row of data frame to list*</br>R
data.frame(a=6:10,b=1:5%each%{"b"%.%.}) %>% dfRowToList
$V1
a b
6 b1
Levels: 6 b1
$V2
a b
7 b2
Levels: 7 b2
$V3
a b
8 b3
Levels: 8 b3
$V4
a b
9 b4
Levels: 9 b4
$V5
a b
10 b5
Levels: 10 b5
**dfToHTML(#data.frame)**</br>
*show an data frame in html (only in jupyter notebook)*</br>
# Aleatoire</br>
Aleatoire</br>
# Categorical</br>
**corrCatCon(#vector,#vector)**</br>
*graph of the values in the first params for each cat in the second parameters*</br>R
iris$Sepal.Length %,% iris$Species %...>% graphCatCon
#SAME AS graphCatCon(iris$Sepal.Length,iris$Species)
#SAME AS iris %$% graphCatCon(Sepal.Length,Species)
```
%by%
create list with as name differents categories and as value, the value associate with this value
```R
iris$Sepal.Length %by% iris$Species
$setosa
[1] 5.1 4.9 4.7 4.6 5.0 5.4 4.6 5.0 4.4 4.9 5.4 4.8 4.8 4.3 5.8 5.7 5.4 5.1 5.7 5.1 5.4
[22] 5.1 4.6 5.1 4.8 5.0 5.0 5.2 5.2 4.7 4.8 5.4 5.2 5.5 4.9 5.0 5.5 4.9 4.4 5.1 5.0 4.5
[43] 4.4 5.0 5.1 4.8 5.1 4.6 5.3 5.0
$versicolor
[1] 7.0 6.4 6.9 5.5 6.5 5.7 6.3 4.9 6.6 5.2 5.0 5.9 6.0 6.1 5.6 6.7 5.6 5.8 6.2 5.6 5.9
[22] 6.1 6.3 6.1 6.4 6.6 6.8 6.7 6.0 5.7 5.5 5.5 5.8 6.0 5.4 6.0 6.7 6.3 5.6 5.5 5.5 6.1
[43] 5.8 5.0 5.6 5.7 5.7 6.2 5.1 5.7
$virginica
[1] 6.3 5.8 7.1 6.3 6.5 7.6 4.9 7.3 6.7 7.2 6.5 6.4 6.8 5.7 5.8 6.4 6.5 7.7 7.7 6.0 6.9
[22] 5.6 7.7 6.3 6.7 7.2 6.2 6.1 6.4 7.2 7.4 7.9 6.4 6.3 6.1 7.7 6.3 6.4 6.0 6.9 6.7 6.9
[43] 5.8 6.8 6.7 6.7 6.3 6.5 6.2 5.9
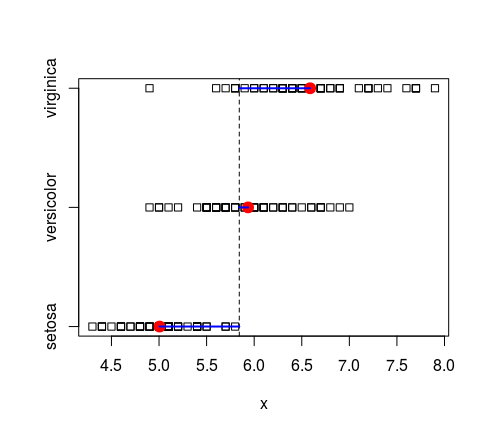
**%byGraph**</br>
*same as %by% but plot graph(see corrCatCon) of values for differents cat (only works if the right arguments is a vector)*</br>R
iris$Sepal.Length %byGraph% iris$Species
$setosa
[1] 5.1 4.9 4.7 4.6 5.0 5.4 4.6 5.0 4.4 4.9 5.4 4.8 4.8 4.3 5.8 5.7 5.4 5.1 5.7 5.1 5.4
[22] 5.1 4.6 5.1 4.8 5.0 5.0 5.2 5.2 4.7 4.8 5.4 5.2 5.5 4.9 5.0 5.5 4.9 4.4 5.1 5.0 4.5
[43] 4.4 5.0 5.1 4.8 5.1 4.6 5.3 5.0
$versicolor
[1] 7.0 6.4 6.9 5.5 6.5 5.7 6.3 4.9 6.6 5.2 5.0 5.9 6.0 6.1 5.6 6.7 5.6 5.8 6.2 5.6 5.9
[22] 6.1 6.3 6.1 6.4 6.6 6.8 6.7 6.0 5.7 5.5 5.5 5.8 6.0 5.4 6.0 6.7 6.3 5.6 5.5 5.5 6.1
[43] 5.8 5.0 5.6 5.7 5.7 6.2 5.1 5.7
$virginica
[1] 6.3 5.8 7.1 6.3 6.5 7.6 4.9 7.3 6.7 7.2 6.5 6.4 6.8 5.7 5.8 6.4 6.5 7.7 7.7 6.0 6.9
[22] 5.6 7.7 6.3 6.7 7.2 6.2 6.1 6.4 7.2 7.4 7.9 6.4 6.3 6.1 7.7 6.3 6.4 6.0 6.9 6.7 6.9
[43] 5.8 6.8 6.7 6.7 6.3 6.5 6.2 5.9

**int.hist**</br>
# Usefull Utils</br>
## capture</br>
**captureCat**</br>
*capture in a variable the result of the cat function for the given parameter*</br>R
a=1:5
a
[1] 1 2 3 4 5
az=captureCat(a)
az
[1] "1 2 3 4 5"
*capturePrint*</br>
*capture in a variable the result of the cat function for the given parameter*</br>R
a=lm(Petal.Width ~ Sepal.Length,data=iris)
print(a)
Call:
lm(formula = Petal.Width ~ Sepal.Length, data = iris)
Coefficients:
(Intercept) Sepal.Length
-3.2002 0.7529
az=capturePrint(a)
az
[1] ""
[2] "Call:"
[3] "lm(formula = Petal.Width ~ Sepal.Length, data = iris)"
[4] ""
[5] "Coefficients:"
[6] " (Intercept) Sepal.Length "
[7] " -3.2002 0.7529 "
[8] ""
## embed</br>
**embed**(x, height="100%",width="100%") </br>
*Display widget x, in html in jupyter notebook*</br>
*(see DT::datatable)*</br></br>R
graph=dataSim %>% ggplot(aes(x=time,y=bin1)) + geom_point() + geom_point_interactive(aes(data_id=rownames(dataSim)), size = 2) + theme_minimal()
graphWidget= graph %>% girafe(ggobj = .) %>% girafe_options(opts_hover(css = "fill:red;r:4pt;"))
embed(graphWidget,"500px","70%")
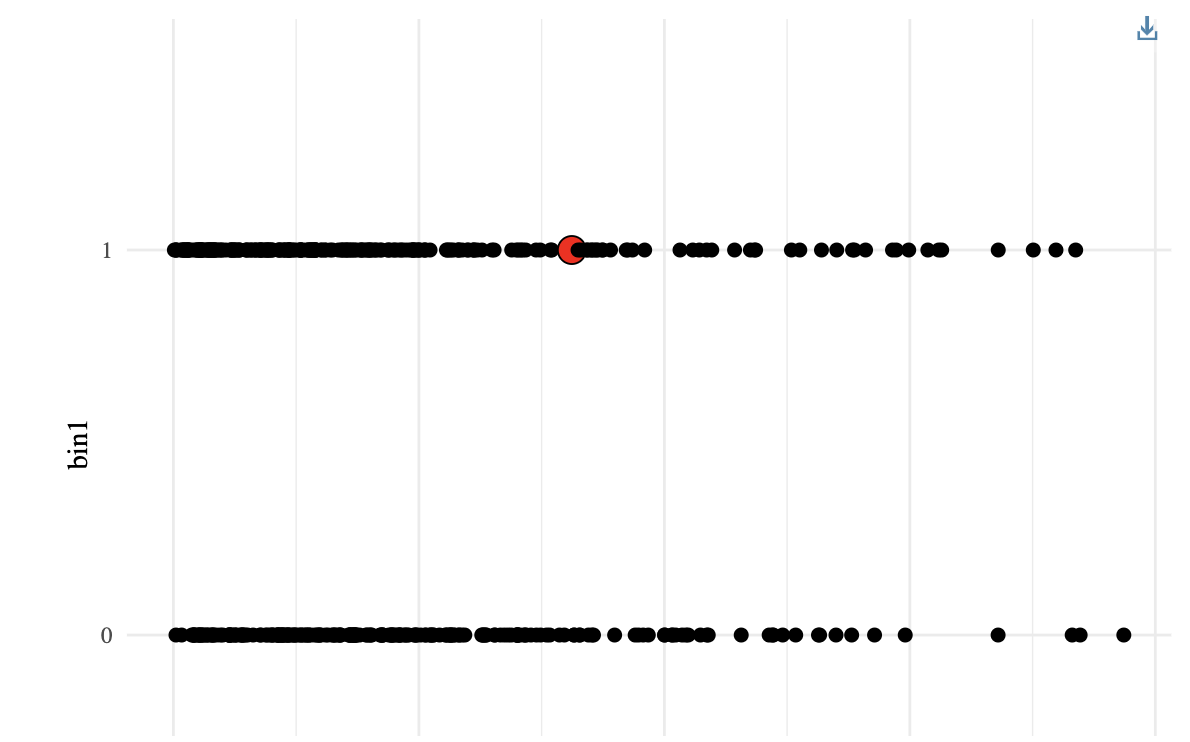
*(see ggirafe)*</br></br>
**embedDT**(dt,height="100%",width="100%",...)</br>
*Display a data frame in html beautiful table interactive in **jupyter notebook***</br>
*DT::datatable(dt,...)*
```R
embedDT(dataSim,"500px","100%",filter="top")
#if Rstudio
DT::datatable(iris,filter="top")
```
*(see embed)*</br>
## plotWH
**plotWH**(w=NULL,h=NULL)-> reset (it is a function)</br>
*Change Plot Width/height*</br>
```R
resetWH = plotWH(w=10)
... #plot graphique
resetWH() # when finish with modified width/height
```
*(options repr.plot.(width/height))*</br>
## Warnings
**showWarning()**</br>
**hideWarning()**</br>
**toggleWarning()**</br>
```R
> testit <- function() warning("testit")
> testit() #Warn
Warning message:
In testit() : testit
> hideWarning()
> testit() #not Warn
> showWarning()
> testit() #Warn
Warning message:
In testit() : testit
> toggleWarning()
> testit() #not Warn
> toggleWarning()
> testit() #Warn
Warning message:
In testit() : testit
```
**tg**(smth)</br>
*hide/suppress warnings and messages*</br>
```R
tg(smthWithWarningsOrMessages)
```
**suppressWarningsGgplot**(ggplotToPlot)</br>
*hide ggplots warnings*
```R
suppressWarningsGgplot(ggplotPlot)
```
## Reduce and Filter
### Reduce
**%reduce%(x,ops)**</br>
*Reduce list x with operator ops*</br>
*(Reduce(x,f=ops))*</br>
```R
> a=1:5
> a %reduce% "+"
[1] 15
```
### Filter
**%filter%(#list,#fn)**</br>
*Filter list with function fn*</br>
```R
> 1:5 %filter% l1__( .%%2==0 ) #SAME AS 1:5 %filter% l1_(~.%%2==0 ) #SAME AS 1:5 %filter% function(.) .%%2==0 #SAME AS 1:5 %filter% lambda(.,.%%2==0) #(see wrapr)
[1] 2 4
```
### Select Cols</br>
**catCol(#dataframe) notCatCol(#dataframe)**</br>
*Select categorical columns in #dataframe*</br>
```R
> iris %>% catCol %>% str
'data.frame': 150 obs. of 1 variable:
$ Species: Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
> iris %>% notCatCol %>% str
'data.frame': 150 obs. of 4 variables:
$ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
$ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
$ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
$ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
```
**numericCol(#dataframe) notNumericCol(#dataframe)**</br>
*Select numerical columns in #dataframe*</br>
```R
> iris %>% numericCol() %>% str
'data.frame': 150 obs. of 4 variables:
$ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
$ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
$ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
$ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
> iris %>% notNumericCol() %>% str
'data.frame': 150 obs. of 1 variable:
$ Species: Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
```
# Usefull plot </br>
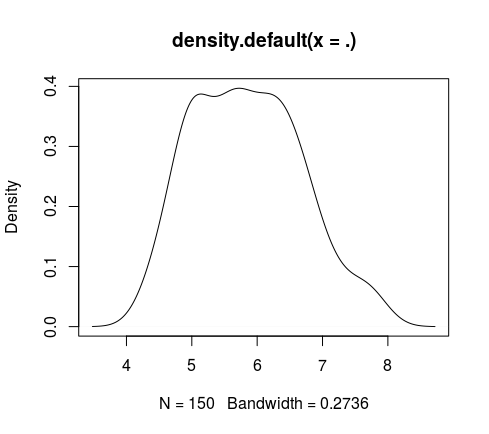
**densityPlt(#numericVector)**</br>
*Give and plot the density of the passed vector (density + plot)(stats::density)(graphics::plot)*</br>
```R
> iris %>% numericCol %getCol% 1 %>% densityPlt
```
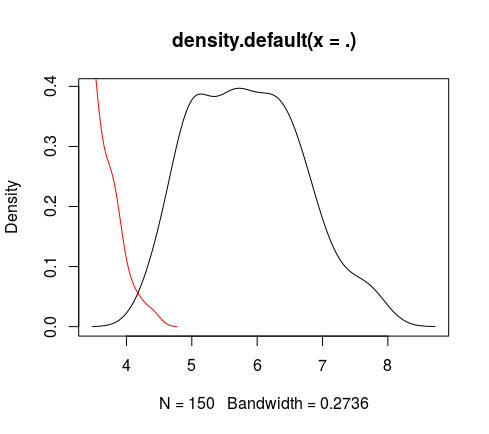
**densityLines(#numericVector)**</br>
*Give and plot the density but here use: lines (graphics::lines) (usefull for append density to an existing plot)*</br>
```R
> iris %>% numericCol %getCol% 1 %>% densityPlt
> iris %>% numericCol %getCol% 2 %>% densityLines(col="red")
#SAME
> iris %>% numericCol %getCol% 1:2 %eachFn% l_(densityPlt,densityLines(col="red")) (by default eachFn use names(), and names for data.frame is colnames)
```
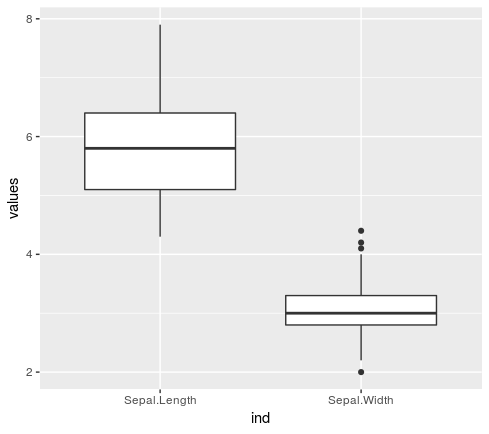
**qplotSameGraphEachCol(#matrix|#data.frame,...)**</br>
*Plot geom asked for each col in d (boxplot,violin,...)*</br>
```R
> iris %>% numericCol %getCol% 1:2 %>% qplotSameGraphEachCol(geom="boxplot")
```
**hidePlot(func)**</br>
*Hide plot printed in func (Exemple with thomasp85/patchwork)*</br>
```R
hidePlot(
{#create plots
numCol=iris %>% numericCol #select numeric cols
b=numCol %>% qplotSameGraphEachCol(geom="boxplot",main="boxplot") #plot boxplot in same graph for each cols
a=numCol %>% as.data.frame %eachCol% {l1_(qplot(.,main=.y))} %reduce% "+" # combine plots
}
)
(#combine plots
b + {
a + plot_layout(1)
} + plot_annotation("Boxplot + hist for Num Var")
) %>%
suppressWarningsGgplot
```
**loadPlotUsefull**</br>
# Usefull Library
**lib + #string|#Stringvector**</br>
*load package or if not exist, install and load it*</br>
```R
lib + "dplyr"
lib + c("purrr","wrapr") + "ggplot2" + c("git:luluperet/icor","git:thomasp85/patchwork","MASS")
lib + "git:cran/MASS"
load same as base::library
# Usefull Packages Icor
reloadIcor()
detach and load icor
update(upgrade=F,...)
Update icor package,(call install_github)
updateReloadIcor(...)
Update icor and reload
detachFast(name#string)
detach package with name
```R
detach("dplyr")
# Each + Map</br>
## Each
**%each(p1#list|#atomic,p2#listFunction|#function)%**</br>
*For each elements in p1 (or juste for the element p1), do each elements in p2 (or just do the element p2) And combine them*</br>
*it's possible to have name with list*</br>R
5 %each% rnorm(20,mean=.)
[,1]
[1,] 5.208568
[2,] 5.241304
[3,] 4.129042
[4,] 5.626426
[5,] 4.788172
[6,] 4.337919
[7,] 4.899254
[8,] 4.749950
[9,] 4.568605
[10,] 4.952485
[11,] 4.458688
[12,] 4.411290
[13,] 3.838677
[14,] 4.330359
[15,] 5.266607
[16,] 4.459932
[17,] 6.676035
[18,] 5.953141
[19,] 5.266492
[20,] 3.583078
5 %each% l_(~rnorm(10,mean=.),~rt(10,df = .))
[,1]
[1,] 2.8300541
[2,] 4.8927280
[3,] 4.7541910
[4,] 4.6632927
[5,] 4.7845754
[6,] 4.3860661
[7,] 6.8552386
[8,] 4.4717009
[9,] 5.9567871
[10,] 5.2502613
[11,] 0.5078755
[12,] -1.2782019
[13,] -1.8228135
[14,] 1.3138229
[15,] 2.3206288
[16,] 0.4834326
[17,] -0.1172201
[18,] -0.1487688
[19,] 2.4639320
[20,] -0.1883499
l(param1=5) %each% l_(~rnorm(5,mean=.),~rt(5,df = .))
[,1]
[1,] 2.8300541
[2,] 4.8927280
[3,] 4.7541910
[4,] 4.6632927
[5,] 4.7845754
[6,] 4.3860661
[7,] 6.8552386
[8,] 4.4717009
[9,] 5.9567871
[10,] 5.2502613
[11,] 0.5078755
[12,] -1.2782019
[13,] -1.8228135
[14,] 1.3138229
[15,] 2.3206288
[16,] 0.4834326
[17,] -0.1172201
[18,] -0.1487688
[19,] 2.4639320
[20,] -0.1883499
l(param1=5) %each% l_(~rnorm(5,mean=.),~rt(5,df = .))
param1
[1,] 4.9572221
[2,] 4.4964496
[3,] 5.2982058
[4,] 4.1527408
[5,] 6.2592159
[6,] 0.4093040
[7,] 0.1006199
[8,] -1.2129520
[9,] -0.1663486
[10,] 0.2167459
l(param1=5,param2=10) %each% l_(normal=~rnorm(10,mean=.),student=~rt(10,df = .))
param1 param2
[1,] 5.7367601 9.42105578
[2,] 6.0204500 11.28830180
[3,] 4.6171627 11.05506386
[4,] 3.6562342 8.31786148
[5,] 3.7550892 10.62510488
[6,] 5.5858563 10.05422571
[7,] 4.2535033 10.03610482
[8,] 3.8561362 9.58243084
[9,] 5.2993138 10.96269036
[10,] 3.8269837 12.08496308
[11,] 0.4619335 0.48020012
[12,] -0.5076719 -1.87017737
[13,] -0.5319049 -2.35908234
[14,] -1.3228157 1.50055514
[15,] -0.1864516 0.34109743
[16,] -1.1957955 0.02759116
[17,] -0.2537597 1.72938623
[18,] -1.6788381 0.65561173
[19,] 1.3222800 -0.08624319
[20,] -0.3316957 -1.05160888
**%each:(p1#list|#atomic,p2#listSpe) (see below)%**</br>
*list spe -> !fn1:fn2:fn3:fn4 each fn will be formula*</br>R
l(param1=5,param2=10) %each:% !rnorm(10,mean=.):rt(10,df=.)
param1 param2
[1,] 3.9852628 10.8618801
[2,] 4.0229170 10.0389925
[3,] 4.5367383 10.0185098
[4,] 4.5252521 9.9040710
[5,] 4.0988606 10.2416906
[6,] 5.2879876 11.2145030
[7,] 4.4056529 11.7918208
[8,] 5.9926545 8.6528593
[9,] 5.8810676 8.5943881
[10,] 3.9937171 12.1794355
[11,] -2.2952650 1.1307431
[12,] 1.2224384 -0.1587621
[13,] -2.3639076 0.7997264
[14,] -0.2115936 -1.1336077
[15,] -1.1214107 -1.3203418
[16,] 1.2084834 -0.1969208
[17,] -0.6580758 0.5846537
[18,] 1.4053799 -1.8358543
[19,] 0.3603988 0.7041107
[20,] -0.1773637 0.7497855
```
%eachCol%(#data.frame|#matrix,#fn) %eachRow%(#data.frame|#matrix,#fn)
for each col/row do function fn
fn = function(.,.y) (.y will be the rownames/colnames) (if the function have a parameter "..." or ".y")
```R
iris %>% numericCol %eachCol% mean
[[1]]
[1] 5.843333
[[2]]
[1] 3.057333
[[3]]
[1] 3.758
[[4]]
[1] 1.199333
iris %>% numericCol %getRow% 1:5 %eachRow% sd
[[1]]
[1] 2.179449
[[2]]
[1] 2.03695
[[3]]
[1] 1.997498
[[4]]
[1] 1.912241
[[5]]
[1] 2.156386
**%eachRowCol.%(#data.frame|#matrix,#fn)**</br>
*apply*</br>
**%eachFn%(p1#list,p2#listOfFn) %eachFnTg%(p1#list,p2#listOfFn)**</br>
*call functions in p2 with the right element in the list p1 (the first element in p1 wille be the parameters of the first function in p2 *</br>
*%eachFnTg% same but don't show result*</br>R
l(10,5) %eachFn% l_(rnorm(10,.),rt(2,.))
[[1]]
[1] 11.322321 9.024418 10.128791 10.043028 9.558132 8.285182 10.534494 10.839975
[9] 8.811831 12.208134
[[2]]
[1] -0.3284563 0.9044645
iris %>% numericCol %getCol% 1:3 %eachCol% density %eachFnTg% l_(plot(main="iris col 1:3"),lines(col="red"),lines(col="yellow"))
## MAP
**%map%**</br>
*like %each% but return a list*</br>
```R
> 5 %map% l_(~rnorm(10,mean=.),~rt(10,df = .))
[[1]]
[[1]][[1]]
[1] 6.217333 7.817015 5.542896 6.197506 4.536185 6.156209 4.676093 4.671299 4.556994
[10] 5.702077
[[1]][[2]]
[1] 1.0574214 0.5550782 -1.2997765 -0.7669661 0.7797546 -0.3401625 1.2158549
[8] -0.8133232 0.2269967 -0.2361220
> l(param1=5) %map% l_(normal=~rnorm(10,mean=.),student=~rt(10,df = .))
$param1
$param1$normal
[1] 4.753331 5.201445 5.393153 4.636426 5.450957 5.222474 5.324348 5.826944 4.682138
[10] 5.813726
$param1$student
[1] -0.44942978 -0.14939527 0.02807041 2.53607663 0.66900864 0.01629510 -0.58214031
[8] -0.03030529 0.43651053 0.92873499
> l(param1=5,param2=10) %map% l_(normal=~rnorm(10,mean=.),student=~rt(10,df = .))
$param1
$param1$normal
[1] 5.032082 7.148801 6.323189 7.157312 3.455346 4.563776 4.200298 4.865634 4.702481
[10] 2.249695
$param1$student
[1] -0.8267602 -0.2121175 0.4930985 -0.3130002 1.5606819 -0.7043366 1.0136329
[8] -0.2031532 -0.8199970 -0.6048155
$param2
$param2$normal
[1] 9.895135 10.596231 9.490120 8.964490 10.597482 11.479018 9.687607 10.212865
[9] 10.504445 10.689903
$param2$student
[1] 0.77245027 -0.51654366 0.54893002 0.84684148 0.41709451 1.84699802 0.17552991
[8] -1.24288775 0.01882849 -1.45396199
```
**%mapFns%**</br>
*invoke_map*</br>
%Xtimes%</br>
# List Sequence Formula Customize</br>
formulatoList.</br>
%from%</br>
# Getter </br>
%getCol% %getRow% %getCol.% %getRow.%</br>
%getElem% %getElem2% %getElems%</br>
# Icor</br>
icor</br>
icor.corrToStudent</br>
icor.critical.r</br>
icor.studentToCorr</br>
icor.graph</br>
# Lists</br>
## LISTS OPS
*%#1,#2%*</br>
* #1 must be constructed with _ and .*</br>
* if you want that the left side must be interprete with "l_", you have to write _ for the left (#1)%</br>
* *1 = _ *</br>
* if you want #2 be normal like "l", just write nothing for #1 *</br>
* #1 = _ *</br>
* #2 = *</br>
* Operator = %_,%*</br>
```R
> rnorm(3) %_,% rnorm(3) #SAME AS l( l1_(rnorm(3)), rnorm(3) )
[[1]]
<icor_list>
function (..., .x = ..1, .y = ..2, . = ..1)
rnorm(., 3)
[[2]]
[1] 0.2080809 1.2826301 -0.4169875
```
* if you want you can change the #2, with "__" for interprete the right side with "l__"*</br>
```R
> rnorm(3) %_,__% rnorm(3) #SAME AS l( l1_(rnorm(3)), l1__(rnorm(3)) )
[[1]]
<icor_list>
function (..., .x = ..1, .y = ..2, . = ..1)
rnorm(., 3)
[[2]]
<icor_list>
function (..., .x = ..1, .y = ..2, . = ..1)
rnorm(3)
```
**l l. lx ln lx. ln. %,% %.,.% %.,% %,.% **</br>
*normal list but understand .()*</br>
```R
> i = 3
> l(.(i),4) #SAME .(i) %,% 4
[[1]]
[1] 3
[[2]]
[1] 4
> l.(.(i),4) #SAME .(i) %.,% 4
Error in .(i) : impossible de trouver la fonction "."
ll
normal list but nested list(list())
l_ lx_ ln_ l1_ %,% %,% %,% %__,% %___,% AND symetrics
normal list but for each parameters return a function | l1_ return the first one | %,%
a parameter: function, formula
```R
l_(
rnorm(1),
~rnorm(4),
{ rnorm(mean=4)},
rnorm
) #SAME rnorm(1) %,% (~rnorm(4)) %,% { rnorm(mean=4) } %,% rnorm
[[1]]
function (..., .x = ..1, .y = ..2, . = ..1)
rnorm(., 1)
[[2]]
function (..., .x = ..1, .y = ..2, . = ..1)
rnorm(4)
[[3]]
function (..., .x = ..1, .y = ..2, . = ..1)
{
rnorm(mean = 4)
}
[[4]]
function (..., .x = ..1, .y = ..2, . = ..1)
rnorm(.)
l1_(
rnorm(1),
~rnorm(4),
{ rnorm(mean=4) },
rnorm
) # SAME AS rnorm(1) %,% (~rnorm(4)) %,% { rnorm(mean=4) } %,% rnorm %...>% l1
function (..., .x = ..1, .y = ..2, . = ..1)
rnorm(., 1)
3 %,_% rnorm(1)
[[1]]
[1] 3
[[2]]
function (..., .x = ..1, .y = ..2, . = ..1)
rnorm(., 1)
```
l__ %__,__% %__,_% %__,% AND symetrics
l___
%listToDotsFn_% %listToDotsFn%
# StrCls
StrCls
?? lapplys ??
# Tests
test_normal
test_same_distrib
# Future
(HenrikBengtsson/future)
%future%
//e= d %>% girafe(ggobj = .) %>% girafe_options(opts_hover(css = "fill:red;r:4pt;"))
luluperet/icor documentation built on May 26, 2019, 6:54 p.m.
ICOR, Correlation and Usefull Functions, Operators and Classes
EXAMPLES
iris %>%
numericCol %getCol%
1:4 %>skip>% {
layout_build_matrix(
1,2,3|
0,4,0
);
resetPlt = plotWH(7,5)
}%each% l__(
{
name=names(dimnames(.))
hist(.x,xlab = name,main = name%.%" hist + Density",probability = T);
densityLines(.x,col="red")
}
) %>% invisible
#SAME
#We use eachCol instead of each (.y is the colname)
#We use tg instead of invisible (same, but remove warnings too)
iris %>%
numericCol %getCol%
1:4 %>skip>% {
layout_build_matrix(
1,2,3|
0,4,0
);
resetPlt = plotWH(7,5)
} %eachCol% l1__(
{
name=.y
hist(.x,xlab = name,main = name%.%" hist + Density",probability = T);
densityLines(.x,col="red")
}
) %>% tg

EQ
Test values each in vector < <= == >= > %||% ```R
c(4,2,1) %||% c(9,4,0) < 3 [1] FALSE
**%|%**</br>R c(4,2,1) %|% c(9,4,0) < 3 [1] FALSE TRUE TRUE**%&&%** </br>R c(4,2,1) %&&% c(9,4,0) < 3 [1] FALSE**%&%**</br>R c(4,2,1) %&% c(9,4,0) < 3 [1] FALSE FALSE TRUE ```
list To Parameters of a Function
Call function of the right side, with elements of the list passed in the left side
Operators
%...>%(#list|#atomic,#function) (keep names) ```R
list(mean=10,n=5,sd = 2) %...>% rnorm [1] 12.817348 10.284119 11.355081 7.372176 7.795275
list(mean=10,n=5,sd = 2) %...>% smth List of 3 $ mean: num 10 $ n : num 5 $ sd : num 2
10 %,% 20 %...>% rnorm [1] 19.51830 19.32193 17.89852 20.27248 19.79469 22.44875 21.06525 22.81541 20.42578 [10] 20.35683
(see #icor_lists below)
10 %...>% rnorm [1] 0.88250520 -1.06436486 0.44685920 -0.58315920 1.05593594 0.28902886 -0.01550811 [8] 0.76758399 2.07238604 -0.86256077
**%..._>%(#list|#atomic,#function) (not keep names)**R
list(mean=10,n=5,sd = 2) %..._>% rnorm [1] 4.5024198 8.4893204 10.0372266 4.5468650 3.7732437 4.1219222 [7] 3.7688349 0.1318678 0.9097222 7.8097455
list(mean=10,n=5,sd = 2) %..._>% smth List of 3 $ : num 10 $ : num 5 $ : num 2
10 %,% 20 %...>% rnorm [1] 20.33436 19.57912 19.15201 18.57136 19.53694 19.09004 18.62865 19.14920 19.08933 [10] 19.00091
#(see #icor_lists below)
10 %..._>% rnorm [1] 0.88250520 -1.06436486 0.44685920 -0.58315920 1.05593594 0.28902886 -0.01550811 [8] 0.76758399 2.07238604 -0.86256077
**%listToDotsFn%(#list,#function) (keep names)**R
list(mean=10,n=5,sd = 2) %listToDotsFn% rnorm [1] 11.396479 10.744122 8.012919 11.297808 12.601390
list(ind="a",day=3,month=4) %listToDotsFn% smth List of 3 $ ind : chr "a" $ day : num 3 $ month: num 4
**%listToDotsFn_%(#list,#function) (not keep names)**
*(see "smth" below)*</br>R
list(ind="a",day=3,month=4) %listToDotsFn_% smth List of 3 $ : chr "a" $ : num 3 $ : num 4
10 %,% 20 %listToDotsFn_% rnorm [1] 20.33436 19.57912 19.15201 18.57136 19.53694 19.09004 18.62865 19.14920 19.08933 [10] 19.00091
(see #icor_lists below)
```
String Concat
%.=% Concat and assign Strings
> a="Hello "
> a %.=% "World"
> cat(a)
Hello World
%.% Concat strings (paste0) ```R
"ll"%.%"kk"%.%"kkd" [1] "llkkkkd" ```
Usefull Utils
curry(fn) (Currying)[https://en.wikipedia.org/wiki/Currying] ```R
a=curry(rnorm(10)) a function (...) rnorm(10, ...) a(10) [1] 9.822035 9.897887 10.289826 9.588149 9.705572 11.554598 [7] 11.485561 8.851399 12.550492 10.740860
**smth**</br> *Return str of list of dots* w/br>R 3 %,% 4 %,% l(d=3) %->% smth List of 3 $ : num 3 $ : num 4 $ :List of 1 ..$ d: num 3smth(1,2,3,4,5,"kkk",rnorm,l1___(rnorm(10))) List of 8 $ : num 1 $ : num 2 $ : num 3 $ : num 4 $ : num 5 $ : chr "kkk" $ :function (n, mean = 0, sd = 1) $ :function (...) ..- attr(, "class")= chr [1:2] "purrr_function_partial" "function" ..- attr(, "body")= language ~(function (n, mean = 0, sd = 1) .Call(C_rnorm, n, mean, sd))(10, ...) .. ..- attr(, ".Environment")= ..- attr(, "fn")= symbol rnorm
**startsWithFromList(list,string)**</br>
*filter list with element who begin with string*</br>R
c("hello","he lo","hehe","bonjour","au revoir") %>% startsWithFromList("he") [1] "hello" "he lo" "hehe"
**addNamesToList(list,names)**</br>
*add names to list*</br>R
c("hello","he lo","hehe","bonjour","au revoir") %>% addNamesToList(1:5) 1 2 3 4 5 "hello" "he lo" "hehe" "bonjour" "au revoir"
c("hello","he lo","hehe","bonjour","au revoir") %>% addNamesToList(1:5 %each% {"d"%.%.}) d1 d2 d3 d4 d5 "hello" "he lo" "hehe" "bonjour" "au revoir"
l("hello","he lo","hehe","bonjour","au revoir") %>% addNamesToList(1:5 %each% {"d"%.%.}) $d1 [1] "hello"
$d2 [1] "he lo"
$d3 [1] "hehe"
$d4 [1] "bonjour"
$d5 [1] "au revoir"
``` nothing(...) return nothing with whatever params
%call%(#fn,#list) *call fn with elements of list as parameters
%>skip>%(data,#fn) doAndSkip %-|skip|->% Execute fn and return data ! all above do the same
Usefull DataFrame
toDF(list) toDFt(list) convert an list to a dataframe with good colnames, rownames ```R
l("hello","he lo","hehe","bonjour","au revoir") %>% addNamesToList(1:5 %each% {"d"%.%.}) %>% toDF [,1] d1 "hello" d2 "he lo" d3 "hehe" d4 "bonjour" d5 "au revoir"
l("hello","he lo","hehe","bonjour","au revoir") %>% addNamesToList(1:5 %each% {"d"%.%.}) %>% toDFt d1 d2 d3 d4 d5 1 hello he lo hehe bonjour au revoir
**add_row_with_name(#data.frame)**</br>
*add row with rowname in a data frame*</br>R
data.frame(a=6:10,b=1:5%each%{"b"%.%.}) a b 1 6 b1 2 7 b2 3 8 b3 4 9 b4 5 10 b5
data.frame(a=6:10,b=1:5%each%{"b"%.%.}) %>% add_row_with_name(a=9,b="b5",name="9") a b 1 6 b1 2 7 b2 3 8 b3 4 9 b4 5 10 b5 9 9 b5
**dfRowToList**</br>
*convert row of data frame to list*</br>R
data.frame(a=6:10,b=1:5%each%{"b"%.%.}) %>% dfRowToList $V1 a b 6 b1 Levels: 6 b1
$V2 a b 7 b2 Levels: 7 b2
$V3 a b 8 b3 Levels: 8 b3
$V4 a b 9 b4 Levels: 9 b4
$V5 a b 10 b5 Levels: 10 b5
**dfToHTML(#data.frame)**</br>
*show an data frame in html (only in jupyter notebook)*</br>

# Aleatoire</br>
Aleatoire</br>
# Categorical</br>
**corrCatCon(#vector,#vector)**</br>
*graph of the values in the first params for each cat in the second parameters*</br>R
iris$Sepal.Length %,% iris$Species %...>% graphCatCon
#SAME AS graphCatCon(iris$Sepal.Length,iris$Species)
#SAME AS iris %$% graphCatCon(Sepal.Length,Species)
```

%by% create list with as name differents categories and as value, the value associate with this value ```R
iris$Sepal.Length %by% iris$Species $setosa [1] 5.1 4.9 4.7 4.6 5.0 5.4 4.6 5.0 4.4 4.9 5.4 4.8 4.8 4.3 5.8 5.7 5.4 5.1 5.7 5.1 5.4 [22] 5.1 4.6 5.1 4.8 5.0 5.0 5.2 5.2 4.7 4.8 5.4 5.2 5.5 4.9 5.0 5.5 4.9 4.4 5.1 5.0 4.5 [43] 4.4 5.0 5.1 4.8 5.1 4.6 5.3 5.0
$versicolor [1] 7.0 6.4 6.9 5.5 6.5 5.7 6.3 4.9 6.6 5.2 5.0 5.9 6.0 6.1 5.6 6.7 5.6 5.8 6.2 5.6 5.9 [22] 6.1 6.3 6.1 6.4 6.6 6.8 6.7 6.0 5.7 5.5 5.5 5.8 6.0 5.4 6.0 6.7 6.3 5.6 5.5 5.5 6.1 [43] 5.8 5.0 5.6 5.7 5.7 6.2 5.1 5.7
$virginica [1] 6.3 5.8 7.1 6.3 6.5 7.6 4.9 7.3 6.7 7.2 6.5 6.4 6.8 5.7 5.8 6.4 6.5 7.7 7.7 6.0 6.9 [22] 5.6 7.7 6.3 6.7 7.2 6.2 6.1 6.4 7.2 7.4 7.9 6.4 6.3 6.1 7.7 6.3 6.4 6.0 6.9 6.7 6.9 [43] 5.8 6.8 6.7 6.7 6.3 6.5 6.2 5.9
**%byGraph**</br>
*same as %by% but plot graph(see corrCatCon) of values for differents cat (only works if the right arguments is a vector)*</br>R
iris$Sepal.Length %byGraph% iris$Species $setosa [1] 5.1 4.9 4.7 4.6 5.0 5.4 4.6 5.0 4.4 4.9 5.4 4.8 4.8 4.3 5.8 5.7 5.4 5.1 5.7 5.1 5.4 [22] 5.1 4.6 5.1 4.8 5.0 5.0 5.2 5.2 4.7 4.8 5.4 5.2 5.5 4.9 5.0 5.5 4.9 4.4 5.1 5.0 4.5 [43] 4.4 5.0 5.1 4.8 5.1 4.6 5.3 5.0
$versicolor [1] 7.0 6.4 6.9 5.5 6.5 5.7 6.3 4.9 6.6 5.2 5.0 5.9 6.0 6.1 5.6 6.7 5.6 5.8 6.2 5.6 5.9 [22] 6.1 6.3 6.1 6.4 6.6 6.8 6.7 6.0 5.7 5.5 5.5 5.8 6.0 5.4 6.0 6.7 6.3 5.6 5.5 5.5 6.1 [43] 5.8 5.0 5.6 5.7 5.7 6.2 5.1 5.7
$virginica [1] 6.3 5.8 7.1 6.3 6.5 7.6 4.9 7.3 6.7 7.2 6.5 6.4 6.8 5.7 5.8 6.4 6.5 7.7 7.7 6.0 6.9 [22] 5.6 7.7 6.3 6.7 7.2 6.2 6.1 6.4 7.2 7.4 7.9 6.4 6.3 6.1 7.7 6.3 6.4 6.0 6.9 6.7 6.9 [43] 5.8 6.8 6.7 6.7 6.3 6.5 6.2 5.9

**int.hist**</br>
# Usefull Utils</br>
## capture</br>
**captureCat**</br>
*capture in a variable the result of the cat function for the given parameter*</br>R
a=1:5 a [1] 1 2 3 4 5 az=captureCat(a) az [1] "1 2 3 4 5"
*capturePrint*</br>
*capture in a variable the result of the cat function for the given parameter*</br>R
a=lm(Petal.Width ~ Sepal.Length,data=iris) print(a)
Call: lm(formula = Petal.Width ~ Sepal.Length, data = iris)
Coefficients: (Intercept) Sepal.Length -3.2002 0.7529
az=capturePrint(a) az [1] "" [2] "Call:" [3] "lm(formula = Petal.Width ~ Sepal.Length, data = iris)" [4] "" [5] "Coefficients:" [6] " (Intercept) Sepal.Length " [7] " -3.2002 0.7529 " [8] ""
## embed</br>
**embed**(x, height="100%",width="100%") </br>
*Display widget x, in html in jupyter notebook*</br>
*(see DT::datatable)*</br></br>R
graph=dataSim %>% ggplot(aes(x=time,y=bin1)) + geom_point() + geom_point_interactive(aes(data_id=rownames(dataSim)), size = 2) + theme_minimal()
graphWidget= graph %>% girafe(ggobj = .) %>% girafe_options(opts_hover(css = "fill:red;r:4pt;"))
embed(graphWidget,"500px","70%")

*(see ggirafe)*</br></br>
**embedDT**(dt,height="100%",width="100%",...)</br>
*Display a data frame in html beautiful table interactive in **jupyter notebook***</br>
*DT::datatable(dt,...)*
```R
embedDT(dataSim,"500px","100%",filter="top")
#if Rstudio
DT::datatable(iris,filter="top")
```

*(see embed)*</br>
## plotWH
**plotWH**(w=NULL,h=NULL)-> reset (it is a function)</br>
*Change Plot Width/height*</br>
```R
resetWH = plotWH(w=10)
... #plot graphique
resetWH() # when finish with modified width/height
```
*(options repr.plot.(width/height))*</br>
## Warnings
**showWarning()**</br>
**hideWarning()**</br>
**toggleWarning()**</br>
```R
> testit <- function() warning("testit")
> testit() #Warn
Warning message:
In testit() : testit
> hideWarning()
> testit() #not Warn
> showWarning()
> testit() #Warn
Warning message:
In testit() : testit
> toggleWarning()
> testit() #not Warn
> toggleWarning()
> testit() #Warn
Warning message:
In testit() : testit
```
**tg**(smth)</br>
*hide/suppress warnings and messages*</br>
```R
tg(smthWithWarningsOrMessages)
```
**suppressWarningsGgplot**(ggplotToPlot)</br>
*hide ggplots warnings*
```R
suppressWarningsGgplot(ggplotPlot)
```
## Reduce and Filter
### Reduce
**%reduce%(x,ops)**</br>
*Reduce list x with operator ops*</br>
*(Reduce(x,f=ops))*</br>
```R
> a=1:5
> a %reduce% "+"
[1] 15
```
### Filter
**%filter%(#list,#fn)**</br>
*Filter list with function fn*</br>
```R
> 1:5 %filter% l1__( .%%2==0 ) #SAME AS 1:5 %filter% l1_(~.%%2==0 ) #SAME AS 1:5 %filter% function(.) .%%2==0 #SAME AS 1:5 %filter% lambda(.,.%%2==0) #(see wrapr)
[1] 2 4
```
### Select Cols</br>
**catCol(#dataframe) notCatCol(#dataframe)**</br>
*Select categorical columns in #dataframe*</br>
```R
> iris %>% catCol %>% str
'data.frame': 150 obs. of 1 variable:
$ Species: Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
> iris %>% notCatCol %>% str
'data.frame': 150 obs. of 4 variables:
$ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
$ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
$ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
$ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
```
**numericCol(#dataframe) notNumericCol(#dataframe)**</br>
*Select numerical columns in #dataframe*</br>
```R
> iris %>% numericCol() %>% str
'data.frame': 150 obs. of 4 variables:
$ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
$ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
$ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
$ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
> iris %>% notNumericCol() %>% str
'data.frame': 150 obs. of 1 variable:
$ Species: Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
```
# Usefull plot </br>
**densityPlt(#numericVector)**</br>
*Give and plot the density of the passed vector (density + plot)(stats::density)(graphics::plot)*</br>
```R
> iris %>% numericCol %getCol% 1 %>% densityPlt
```

**densityLines(#numericVector)**</br>
*Give and plot the density but here use: lines (graphics::lines) (usefull for append density to an existing plot)*</br>
```R
> iris %>% numericCol %getCol% 1 %>% densityPlt
> iris %>% numericCol %getCol% 2 %>% densityLines(col="red")
#SAME
> iris %>% numericCol %getCol% 1:2 %eachFn% l_(densityPlt,densityLines(col="red")) (by default eachFn use names(), and names for data.frame is colnames)
```

**qplotSameGraphEachCol(#matrix|#data.frame,...)**</br>
*Plot geom asked for each col in d (boxplot,violin,...)*</br>
```R
> iris %>% numericCol %getCol% 1:2 %>% qplotSameGraphEachCol(geom="boxplot")
```

**hidePlot(func)**</br>
*Hide plot printed in func (Exemple with thomasp85/patchwork)*</br>
```R
hidePlot(
{#create plots
numCol=iris %>% numericCol #select numeric cols
b=numCol %>% qplotSameGraphEachCol(geom="boxplot",main="boxplot") #plot boxplot in same graph for each cols
a=numCol %>% as.data.frame %eachCol% {l1_(qplot(.,main=.y))} %reduce% "+" # combine plots
}
)
(#combine plots
b + {
a + plot_layout(1)
} + plot_annotation("Boxplot + hist for Num Var")
) %>%
suppressWarningsGgplot
```

**loadPlotUsefull**</br>
# Usefull Library
**lib + #string|#Stringvector**</br>
*load package or if not exist, install and load it*</br>
```R
lib + "dplyr"
lib + c("purrr","wrapr") + "ggplot2" + c("git:luluperet/icor","git:thomasp85/patchwork","MASS")
lib + "git:cran/MASS"
load same as base::library # Usefull Packages Icor reloadIcor() detach and load icor update(upgrade=F,...) Update icor package,(call install_github) updateReloadIcor(...) Update icor and reload detachFast(name#string) detach package with name ```R detach("dplyr")
# Each + Map</br>
## Each
**%each(p1#list|#atomic,p2#listFunction|#function)%**</br>
*For each elements in p1 (or juste for the element p1), do each elements in p2 (or just do the element p2) And combine them*</br>
*it's possible to have name with list*</br>R
5 %each% rnorm(20,mean=.) [,1] [1,] 5.208568 [2,] 5.241304 [3,] 4.129042 [4,] 5.626426 [5,] 4.788172 [6,] 4.337919 [7,] 4.899254 [8,] 4.749950 [9,] 4.568605 [10,] 4.952485 [11,] 4.458688 [12,] 4.411290 [13,] 3.838677 [14,] 4.330359 [15,] 5.266607 [16,] 4.459932 [17,] 6.676035 [18,] 5.953141 [19,] 5.266492 [20,] 3.583078
5 %each% l_(~rnorm(10,mean=.),~rt(10,df = .)) [,1] [1,] 2.8300541 [2,] 4.8927280 [3,] 4.7541910 [4,] 4.6632927 [5,] 4.7845754 [6,] 4.3860661 [7,] 6.8552386 [8,] 4.4717009 [9,] 5.9567871 [10,] 5.2502613 [11,] 0.5078755 [12,] -1.2782019 [13,] -1.8228135 [14,] 1.3138229 [15,] 2.3206288 [16,] 0.4834326 [17,] -0.1172201 [18,] -0.1487688 [19,] 2.4639320 [20,] -0.1883499
l(param1=5) %each% l_(~rnorm(5,mean=.),~rt(5,df = .)) [,1] [1,] 2.8300541 [2,] 4.8927280 [3,] 4.7541910 [4,] 4.6632927 [5,] 4.7845754 [6,] 4.3860661 [7,] 6.8552386 [8,] 4.4717009 [9,] 5.9567871 [10,] 5.2502613 [11,] 0.5078755 [12,] -1.2782019 [13,] -1.8228135 [14,] 1.3138229 [15,] 2.3206288 [16,] 0.4834326 [17,] -0.1172201 [18,] -0.1487688 [19,] 2.4639320 [20,] -0.1883499
l(param1=5) %each% l_(~rnorm(5,mean=.),~rt(5,df = .)) param1 [1,] 4.9572221 [2,] 4.4964496 [3,] 5.2982058 [4,] 4.1527408 [5,] 6.2592159 [6,] 0.4093040 [7,] 0.1006199 [8,] -1.2129520 [9,] -0.1663486 [10,] 0.2167459
l(param1=5,param2=10) %each% l_(normal=~rnorm(10,mean=.),student=~rt(10,df = .)) param1 param2 [1,] 5.7367601 9.42105578 [2,] 6.0204500 11.28830180 [3,] 4.6171627 11.05506386 [4,] 3.6562342 8.31786148 [5,] 3.7550892 10.62510488 [6,] 5.5858563 10.05422571 [7,] 4.2535033 10.03610482 [8,] 3.8561362 9.58243084 [9,] 5.2993138 10.96269036 [10,] 3.8269837 12.08496308 [11,] 0.4619335 0.48020012 [12,] -0.5076719 -1.87017737 [13,] -0.5319049 -2.35908234 [14,] -1.3228157 1.50055514 [15,] -0.1864516 0.34109743 [16,] -1.1957955 0.02759116 [17,] -0.2537597 1.72938623 [18,] -1.6788381 0.65561173 [19,] 1.3222800 -0.08624319 [20,] -0.3316957 -1.05160888
**%each:(p1#list|#atomic,p2#listSpe) (see below)%**</br>
*list spe -> !fn1:fn2:fn3:fn4 each fn will be formula*</br>R
l(param1=5,param2=10) %each:% !rnorm(10,mean=.):rt(10,df=.) param1 param2 [1,] 3.9852628 10.8618801 [2,] 4.0229170 10.0389925 [3,] 4.5367383 10.0185098 [4,] 4.5252521 9.9040710 [5,] 4.0988606 10.2416906 [6,] 5.2879876 11.2145030 [7,] 4.4056529 11.7918208 [8,] 5.9926545 8.6528593 [9,] 5.8810676 8.5943881 [10,] 3.9937171 12.1794355 [11,] -2.2952650 1.1307431 [12,] 1.2224384 -0.1587621 [13,] -2.3639076 0.7997264 [14,] -0.2115936 -1.1336077 [15,] -1.1214107 -1.3203418 [16,] 1.2084834 -0.1969208 [17,] -0.6580758 0.5846537 [18,] 1.4053799 -1.8358543 [19,] 0.3603988 0.7041107 [20,] -0.1773637 0.7497855
```
%eachCol%(#data.frame|#matrix,#fn) %eachRow%(#data.frame|#matrix,#fn) for each col/row do function fn fn = function(.,.y) (.y will be the rownames/colnames) (if the function have a parameter "..." or ".y") ```R
iris %>% numericCol %eachCol% mean [[1]] [1] 5.843333
[[2]] [1] 3.057333
[[3]] [1] 3.758
[[4]] [1] 1.199333
iris %>% numericCol %getRow% 1:5 %eachRow% sd [[1]] [1] 2.179449
[[2]] [1] 2.03695
[[3]] [1] 1.997498
[[4]] [1] 1.912241
[[5]] [1] 2.156386
**%eachRowCol.%(#data.frame|#matrix,#fn)**</br>
*apply*</br>
**%eachFn%(p1#list,p2#listOfFn) %eachFnTg%(p1#list,p2#listOfFn)**</br>
*call functions in p2 with the right element in the list p1 (the first element in p1 wille be the parameters of the first function in p2 *</br>
*%eachFnTg% same but don't show result*</br>R
l(10,5) %eachFn% l_(rnorm(10,.),rt(2,.)) [[1]] [1] 11.322321 9.024418 10.128791 10.043028 9.558132 8.285182 10.534494 10.839975 [9] 8.811831 12.208134
[[2]] [1] -0.3284563 0.9044645
iris %>% numericCol %getCol% 1:3 %eachCol% density %eachFnTg% l_(plot(main="iris col 1:3"),lines(col="red"),lines(col="yellow"))

## MAP
**%map%**</br>
*like %each% but return a list*</br>
```R
> 5 %map% l_(~rnorm(10,mean=.),~rt(10,df = .))
[[1]]
[[1]][[1]]
[1] 6.217333 7.817015 5.542896 6.197506 4.536185 6.156209 4.676093 4.671299 4.556994
[10] 5.702077
[[1]][[2]]
[1] 1.0574214 0.5550782 -1.2997765 -0.7669661 0.7797546 -0.3401625 1.2158549
[8] -0.8133232 0.2269967 -0.2361220
> l(param1=5) %map% l_(normal=~rnorm(10,mean=.),student=~rt(10,df = .))
$param1
$param1$normal
[1] 4.753331 5.201445 5.393153 4.636426 5.450957 5.222474 5.324348 5.826944 4.682138
[10] 5.813726
$param1$student
[1] -0.44942978 -0.14939527 0.02807041 2.53607663 0.66900864 0.01629510 -0.58214031
[8] -0.03030529 0.43651053 0.92873499
> l(param1=5,param2=10) %map% l_(normal=~rnorm(10,mean=.),student=~rt(10,df = .))
$param1
$param1$normal
[1] 5.032082 7.148801 6.323189 7.157312 3.455346 4.563776 4.200298 4.865634 4.702481
[10] 2.249695
$param1$student
[1] -0.8267602 -0.2121175 0.4930985 -0.3130002 1.5606819 -0.7043366 1.0136329
[8] -0.2031532 -0.8199970 -0.6048155
$param2
$param2$normal
[1] 9.895135 10.596231 9.490120 8.964490 10.597482 11.479018 9.687607 10.212865
[9] 10.504445 10.689903
$param2$student
[1] 0.77245027 -0.51654366 0.54893002 0.84684148 0.41709451 1.84699802 0.17552991
[8] -1.24288775 0.01882849 -1.45396199
```
**%mapFns%**</br>
*invoke_map*</br>
%Xtimes%</br>
# List Sequence Formula Customize</br>
formulatoList.</br>
%from%</br>
# Getter </br>
%getCol% %getRow% %getCol.% %getRow.%</br>
%getElem% %getElem2% %getElems%</br>
# Icor</br>
icor</br>
icor.corrToStudent</br>
icor.critical.r</br>
icor.studentToCorr</br>
icor.graph</br>
# Lists</br>
## LISTS OPS
*%#1,#2%*</br>
* #1 must be constructed with _ and .*</br>
* if you want that the left side must be interprete with "l_", you have to write _ for the left (#1)%</br>
* *1 = _ *</br>
* if you want #2 be normal like "l", just write nothing for #1 *</br>
* #1 = _ *</br>
* #2 = *</br>
* Operator = %_,%*</br>
```R
> rnorm(3) %_,% rnorm(3) #SAME AS l( l1_(rnorm(3)), rnorm(3) )
[[1]]
<icor_list>
function (..., .x = ..1, .y = ..2, . = ..1)
rnorm(., 3)
[[2]]
[1] 0.2080809 1.2826301 -0.4169875
```
* if you want you can change the #2, with "__" for interprete the right side with "l__"*</br>
```R
> rnorm(3) %_,__% rnorm(3) #SAME AS l( l1_(rnorm(3)), l1__(rnorm(3)) )
[[1]]
<icor_list>
function (..., .x = ..1, .y = ..2, . = ..1)
rnorm(., 3)
[[2]]
<icor_list>
function (..., .x = ..1, .y = ..2, . = ..1)
rnorm(3)
```
**l l. lx ln lx. ln. %,% %.,.% %.,% %,.% **</br>
*normal list but understand .()*</br>
```R
> i = 3
> l(.(i),4) #SAME .(i) %,% 4
[[1]]
[1] 3
[[2]]
[1] 4
> l.(.(i),4) #SAME .(i) %.,% 4
Error in .(i) : impossible de trouver la fonction "."
ll normal list but nested list(list()) l_ lx_ ln_ l1_ %,% %,% %,% %__,% %___,% AND symetrics normal list but for each parameters return a function | l1_ return the first one | %,% a parameter: function, formula ```R
l_( rnorm(1), ~rnorm(4), { rnorm(mean=4)}, rnorm ) #SAME rnorm(1) %,% (~rnorm(4)) %,% { rnorm(mean=4) } %,% rnorm [[1]] function (..., .x = ..1, .y = ..2, . = ..1) rnorm(., 1)
[[2]] function (..., .x = ..1, .y = ..2, . = ..1) rnorm(4)
[[3]] function (..., .x = ..1, .y = ..2, . = ..1) { rnorm(mean = 4) }
[[4]] function (..., .x = ..1, .y = ..2, . = ..1) rnorm(.)
l1_( rnorm(1), ~rnorm(4), { rnorm(mean=4) }, rnorm ) # SAME AS rnorm(1) %,% (~rnorm(4)) %,% { rnorm(mean=4) } %,% rnorm %...>% l1 function (..., .x = ..1, .y = ..2, . = ..1) rnorm(., 1)
3 %,_% rnorm(1) [[1]] [1] 3
[[2]] function (..., .x = ..1, .y = ..2, . = ..1) rnorm(., 1)
``` l__ %__,__% %__,_% %__,% AND symetrics l___ %listToDotsFn_% %listToDotsFn% # StrCls StrCls ?? lapplys ?? # Tests test_normal test_same_distrib # Future (HenrikBengtsson/future) %future% //e= d %>% girafe(ggobj = .) %>% girafe_options(opts_hover(css = "fill:red;r:4pt;"))
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
