README.md
In mosscoder/paint2train: What the Package Does (One Line, Title Case)
paint2train package

Background
The purpose of paint2train package is to rapidly label imagery and
spatial data at the pixel level. These labels may in turn be used to
train machine learning algorithms for tasks such as image segmentation.
There are currently four primary functions:
- Generate tiles from larger contiguous sources of multi-spectral
imagery or other spatial data
- Pre-process tiles, generating normalized difference, edge detection,
and neighborhood summary stats layers
- Reduce n layers from pre-processing steps into 3 layers, using UMAP
dimension reduction methods
- Run a local Shiny app to rapidly label pixels based on dissimilarity
thresholds (in UMAP space) to clicked points (labels are saved as
.tifs as you work)
Try a demo of the app!
Installation
The package may be installed via github.
devtools::install_github('mosscoder/paint2train')
Preparatory Steps
Begin by downloading a sample 4-band image.
library(paint2train)
library(ranger)
image_dir <- tempfile()
URL <- 'https://github.com/mosscoder//pt2_supporting_data/blob/main/sample_4band.tif?raw=true'
download.file(url = URL, destfile = image_dir)
par(mfrow = c(2,1))
plotRGB(stack(image_dir)[[1:3]], main = 'True color')
plotRGB(stack(image_dir)[[c(4,2,3)]], main = 'NIR false color')
mtext("15cm true and NIR false color imagery", side = 3, line = -1, outer = TRUE)
par(mfrow = c(1,1))

Next build the directories necessary to house tiles, pre-processed
intermediaries, and labeled data.
tdir <- tempdir()
setwd(tdir) #where output directories will go
preproc_dir <- 'preproc_tiles' #dir for preprocessed tiles
umap_dir <- 'umap_tiles' #dir for UMAP output
lab_dir <- 'label_tiles' #dir for labeled .tif
pred_dir <- 'pred_dir' #a place to predict to new data
lapply(FUN = function(x){dir.create(x)},
X = c(preproc_dir, umap_dir, lab_dir, pred_dir))
Tiling and Pre-processing
Define a 2-column matrix with coordinates corresponding to centroids at
which to generate tiles. Specify tile size and a buffer to avoid edge
effects during pre-processing neighborhood calculations. Specify the
number of cores used during preprocessing. Note that parallel processing
will only work on Unix systems for the native functions in paint2train
at this time. Parallel processing in external functions, such as umap,
will likely work on Windows systems and greatly speed up the dimension
reduction step.
#some test coordinates
xcoords <- c(727495,
727919)
ycoords <- c(5175339,
5175408)
#bind into matrix
coord_mat <- cbind(xcoords, ycoords)
ls <- 30 #how big should the tiles be, this is the side length (in units of data, meters here)
buff <- 5 #buffer in units of data
pre_cores <- ifelse(.Platform$OS.type == 'unix', #how many cores to use for pre-processing
detectCores() - 1,
1)
umap_cores <- detectCores() - 1 #how many cores to use for UMAP dimension reduction
#make 30m tiles with 5m buffer (to avoid edge effects during pre-processing)
tile_at_coords(coords = coord_mat,
len_side = ls,
buffer = buff,
out_dir = preproc_dir,
img = image_dir,
ncores = pre_cores)
 Generate NDVI…
Generate NDVI…
mclapply(
FUN = ndvi_msavi,
X = list.files(preproc_dir, full.names = T),
mc.cores = pre_cores
)
 … and MSAVI values at tiles.
… and MSAVI values at tiles.
 Create edge detection layers by applying a Sobel filter across the first
three PCA axes of data generated thus far.
Create edge detection layers by applying a Sobel filter across the first
three PCA axes of data generated thus far.
mclapply(
FUN = sobel,
X = list.files(preproc_dir, full.names = T),
mc.cores = pre_cores
)
 Calculate mean and variance in 0.25, 0.5, and 1 meter neighborhoods for
first three PCA axes of data generated up to this point. The extents of
these neighborhoods are a critical tuning parameter that will need to be
paired with a particular dataset and modeling objective.
Calculate mean and variance in 0.25, 0.5, and 1 meter neighborhoods for
first three PCA axes of data generated up to this point. The extents of
these neighborhoods are a critical tuning parameter that will need to be
paired with a particular dataset and modeling objective.
neighborhoods <- c(0.25, 0.5, 1) #neighborhood radii in units of imagery
mclapply(
FUN = mean_var,
X = list.files(preproc_dir, full.names = T),
f_width = neighborhoods,
mc.cores = pre_cores
)
As a final pre-processing step, remove the buffers from around each
tile.
mclapply(FUN = remove_buffer,
X = list.files(preproc_dir, full.names = T),
b = buff,
mc.cores = pre_cores)
Here is a sample of the outputs of the focal calculations described
above for the first tile.

Dimension Reduction
Next reduce the pre-processed layers into three dimension with the UMAP
algorithm. This will facilitate identification of similar pixels in the
painting app by conducting non-linear transformations of the data and
mitigate the curse of
dimensionality.
For details on this method, please refer to the UWOT
documentation.
lapply(FUN = umap_tile,
X = list.files(preproc_dir, full.names = TRUE),
out_dir = umap_dir,
n_threads = umap_cores,
n_sgd_threads = umap_cores)
Compare the original RGB tiles with outcomes from pre-processing and
dimension reduction. UMAP space is represented in similarity colors.

Running App Locally
Now we may label our data using the paint2train app. We first define the
classes to label, assigning them an integer value, as well as a
corresponding color palette to visualize labeled areas.
label_key <- list(Unknown = 0,
`Not woody` = 1,
`Woody` = 2)
pal <- c('royalblue',
'tan',
'green')
Provide these lists, location of the UMAP output, and label directory to
the p2t function.
p2t(umap_dir = umap_dir,
label_dir = lab_dir,
label_key = label_key,
label_col = pal)
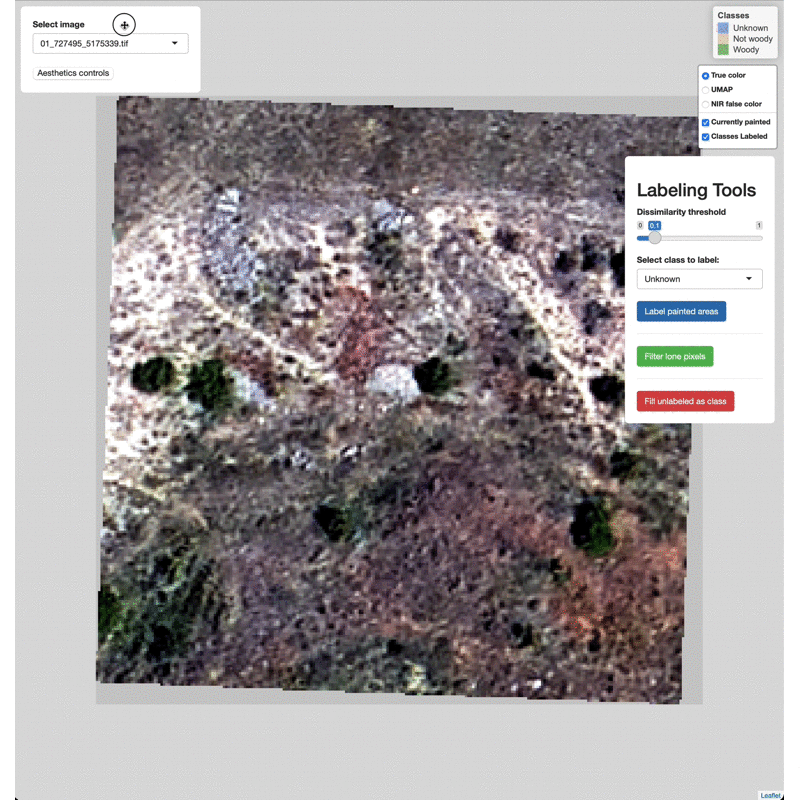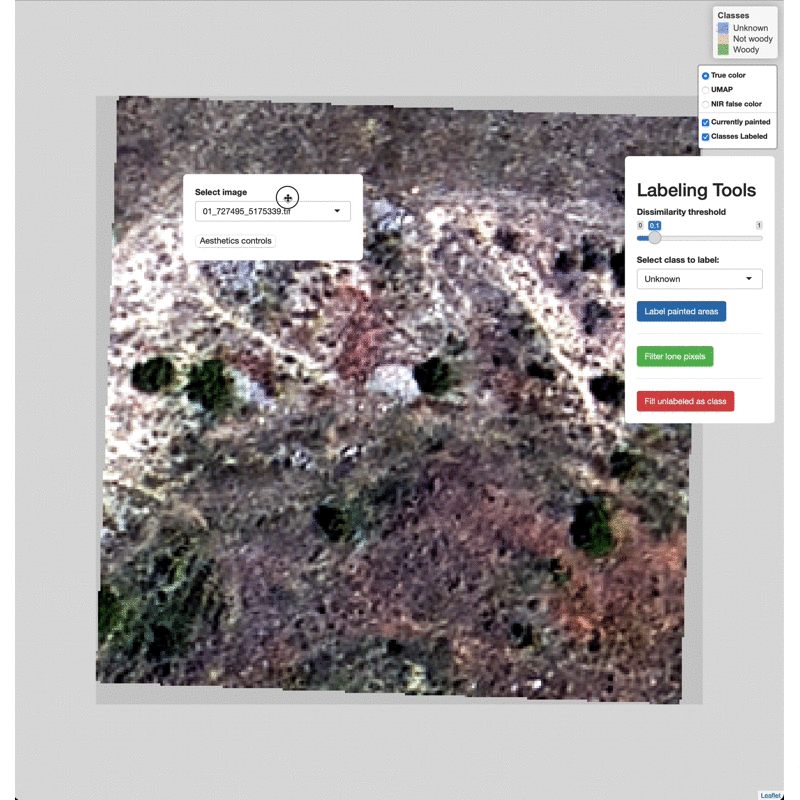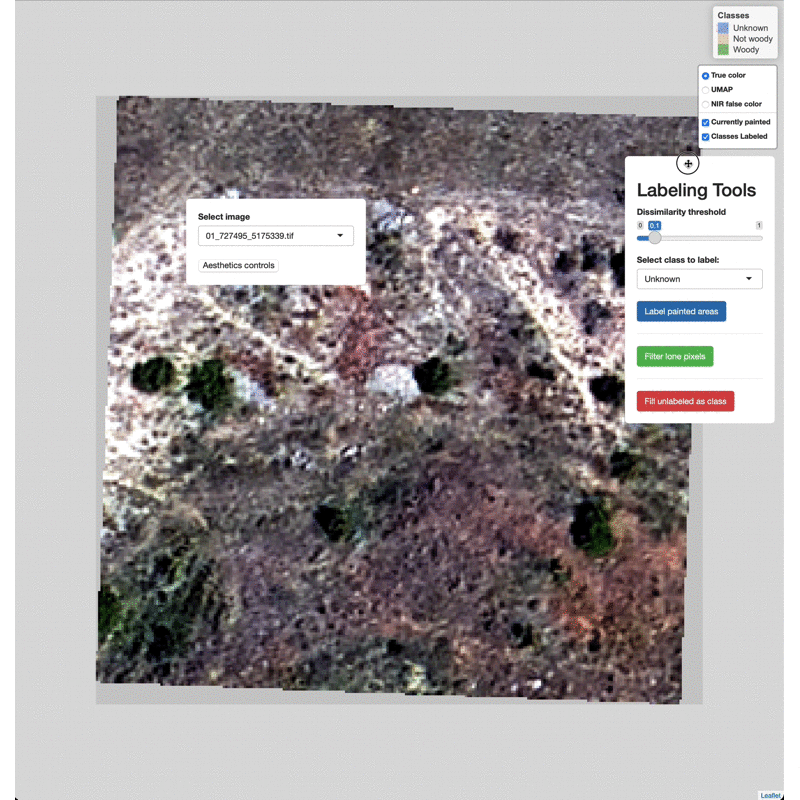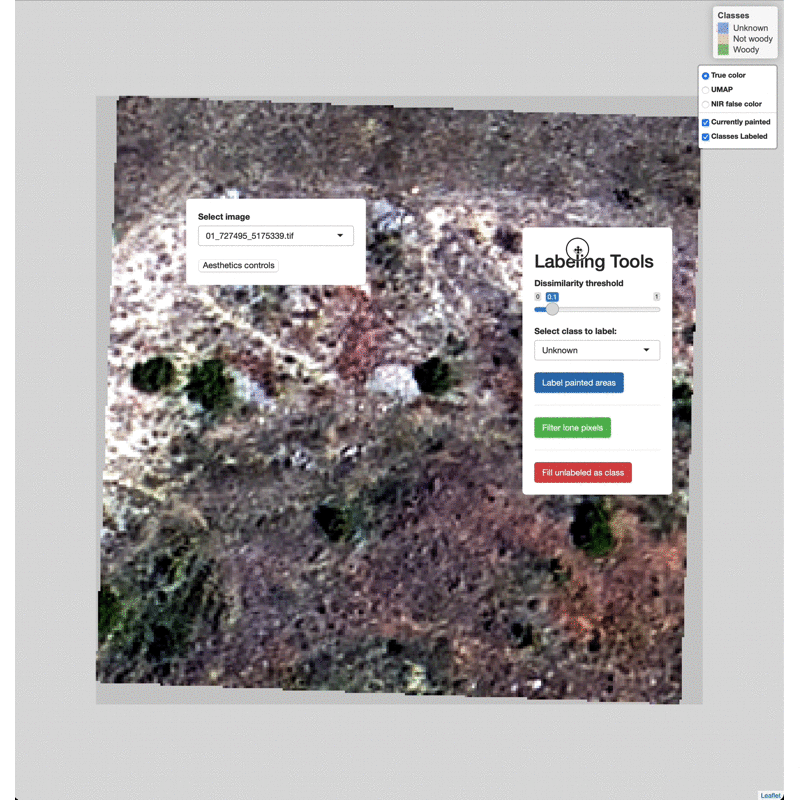
Data Labeling Process
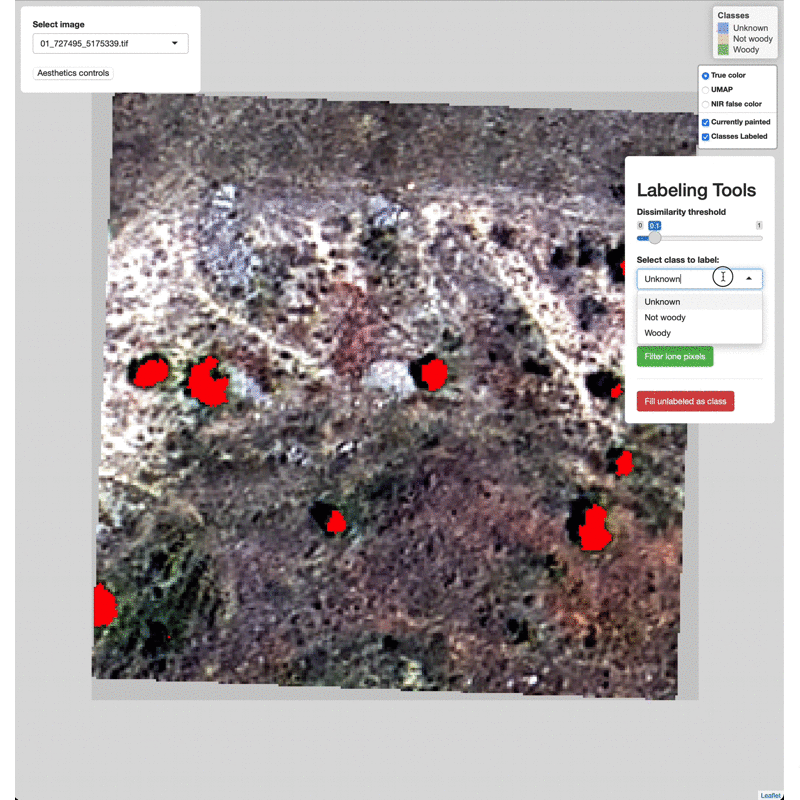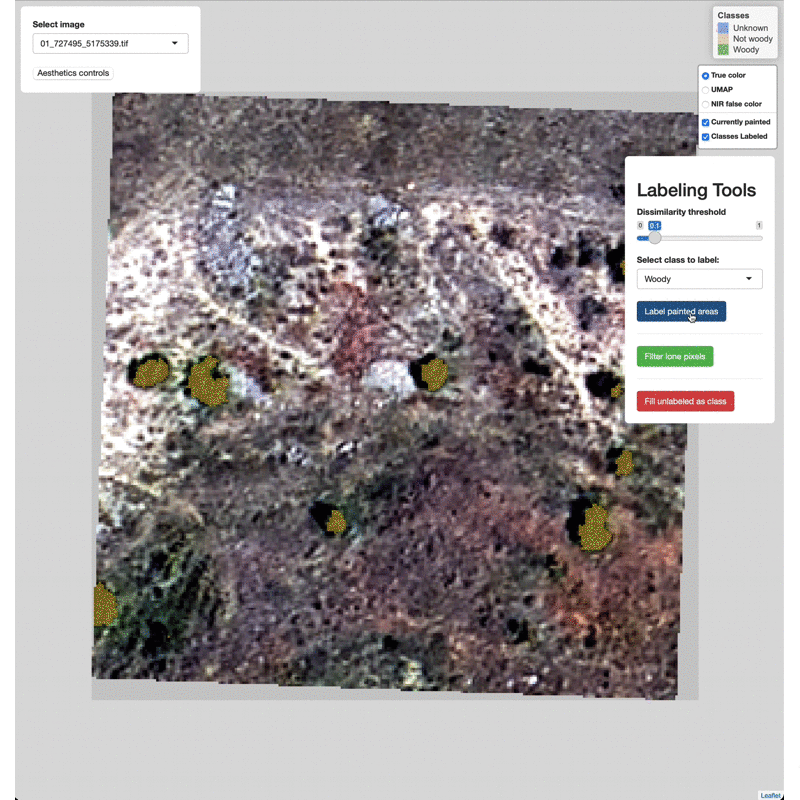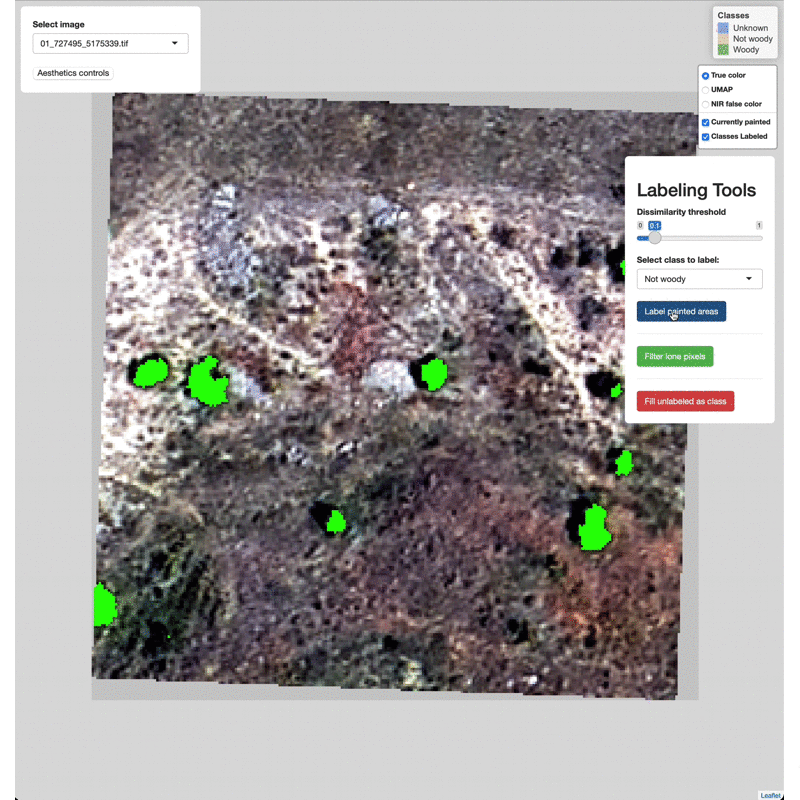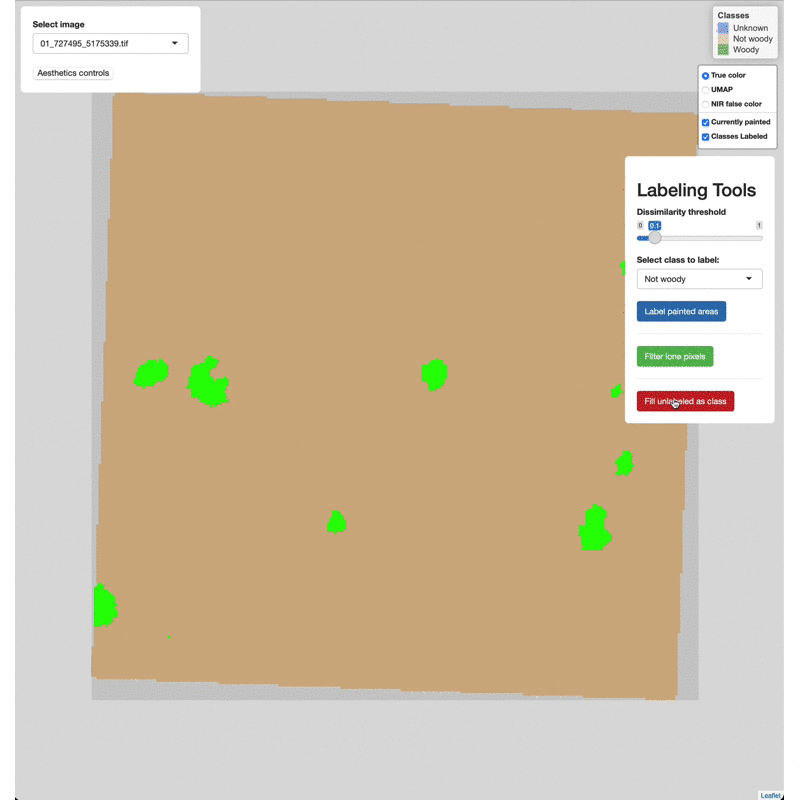
Select imagery tiles from the dropdown menu found in the upper left.

Click on a region you wish to classify and adjust the Dissimilarity
Threshold to match the extent of the class to label.

Select which class to label from the Labeling Tools menu, then click
the Label painted areas button to save the painted pixels to that
class. After painting and labeling focal areas, fill the remaining
unlabeled points by click the FIll unlabeled as class button.
Adjust the color of painted areas from the Aesthetics Controls drop
down menu.

Manually edit pixels by using the draw tools (lower right). Draw a box
or polygon around the region you wish to edit, select the appropriate
class from the Select class to label menu, then click Label drawn
areas
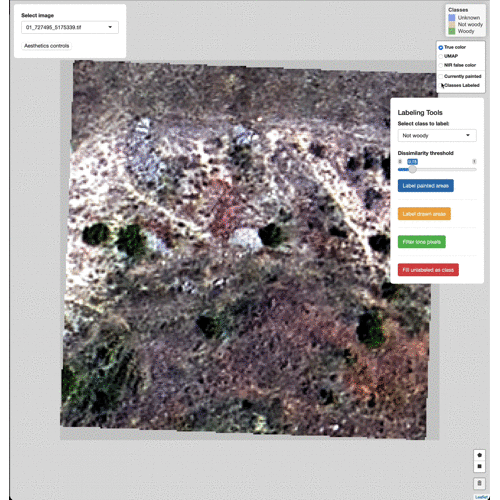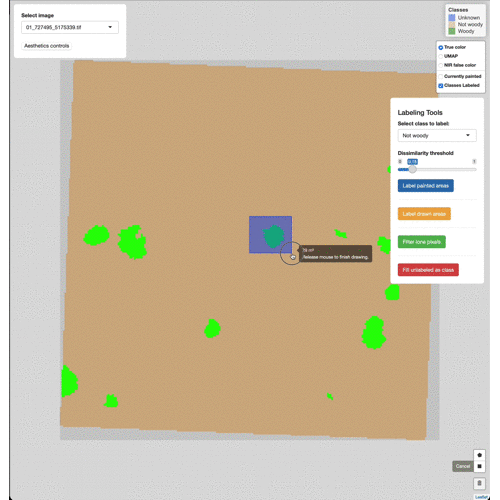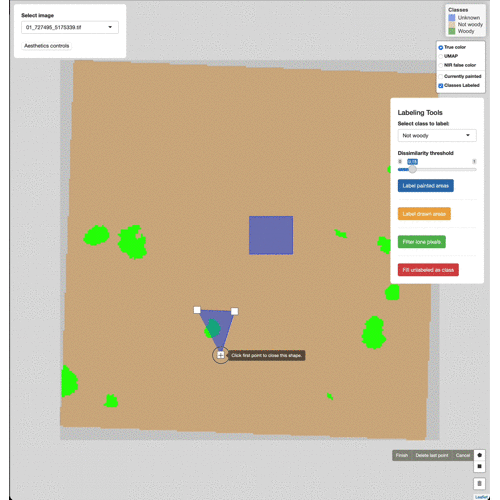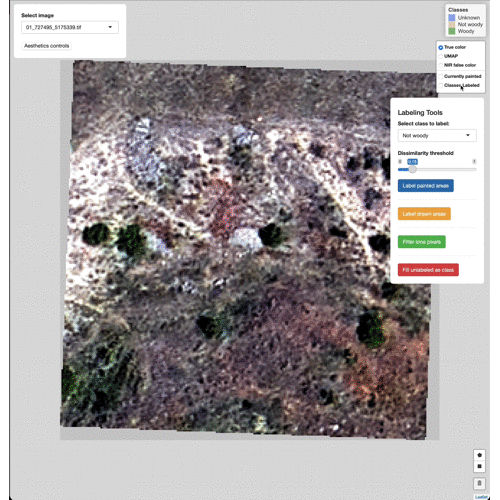
Change the base imagery with the controls in the upper right.

Filter high and low value outlier pixels to brighten or darken base
imagery layers by adjusting the Baselayer quantiles in the
Aesthetics controls drop down menu.

Click and drag to move the controls as needed.
Model Training
Now generate a simple random forest model using the data labeled with
p2t.
train_dat <- load_tdat(preproc_dir = preproc_dir,
label_dir = lab_dir,
ncores = pre_cores)
set.seed(123)
rf_mod <- ranger(label ~ .,
data = train_dat,
num.threads = umap_cores,
classification = TRUE)
print(rf_mod)
#Predict to some new data
og_ext <- extent(stack(image_dir))
mean_x <- mean(og_ext[1:2])
mean_y <- mean(og_ext[3:4])
tile_at_coords(coords = cbind(mean_x, mean_y - 25),
len_side = 25,
buffer = buff,
out_dir = pred_dir,
img = image_dir,
ncores = pre_cores)
pred_files <- list.files(pred_dir, full.names = TRUE)
pre_pipeline <- function(x, fs, b) {
ndvi_msavi(x)
sobel(x)
mean_var(x, f_width = fs)
remove_buffer(x, b)
}
mclapply(FUN = pre_pipeline,
X = pred_files,
mc.cores = pre_cores,
fs = neighborhoods,
b = buff)
pred_names <- colnames(train_dat[2:ncol(train_dat)])
new_dat <- do.call(rbind, mclapply(FUN =
function(x,p){setNames(as.data.frame(getValues(stack(x))), p)},
X = pred_files,
p = pred_names,
mc.cores = pre_cores))
preds <- predict(rf_mod, new_dat, num.threads = umap_cores)
pred_ras <- raster(pred_files[1])
values(pred_ras) <- preds$predictions
leaflet() %>%
addRasterRGB(stack(pred_files[1]),
r = 1, g = 2, b = 3,
group = 'RGB') %>%
addRasterImage(pred_ras,
project = FALSE,
opacity = 0.6,
colors = c('transparent','red'),
group = 'Canopy') %>%
addLayersControl(overlayGroups = c('Canopy'),
options = layersControlOptions(collapsed = FALSE))

mosscoder/paint2train documentation built on Jan. 21, 2022, 11 a.m.
paint2train package

Background
The purpose of paint2train package is to rapidly label imagery and spatial data at the pixel level. These labels may in turn be used to train machine learning algorithms for tasks such as image segmentation.
There are currently four primary functions:
- Generate tiles from larger contiguous sources of multi-spectral imagery or other spatial data
- Pre-process tiles, generating normalized difference, edge detection, and neighborhood summary stats layers
- Reduce n layers from pre-processing steps into 3 layers, using UMAP dimension reduction methods
- Run a local Shiny app to rapidly label pixels based on dissimilarity thresholds (in UMAP space) to clicked points (labels are saved as .tifs as you work)
Try a demo of the app!
Installation
The package may be installed via github.
devtools::install_github('mosscoder/paint2train')
Preparatory Steps
Begin by downloading a sample 4-band image.
library(paint2train)
library(ranger)
image_dir <- tempfile()
URL <- 'https://github.com/mosscoder//pt2_supporting_data/blob/main/sample_4band.tif?raw=true'
download.file(url = URL, destfile = image_dir)
par(mfrow = c(2,1))
plotRGB(stack(image_dir)[[1:3]], main = 'True color')
plotRGB(stack(image_dir)[[c(4,2,3)]], main = 'NIR false color')
mtext("15cm true and NIR false color imagery", side = 3, line = -1, outer = TRUE)
par(mfrow = c(1,1))

Next build the directories necessary to house tiles, pre-processed intermediaries, and labeled data.
tdir <- tempdir()
setwd(tdir) #where output directories will go
preproc_dir <- 'preproc_tiles' #dir for preprocessed tiles
umap_dir <- 'umap_tiles' #dir for UMAP output
lab_dir <- 'label_tiles' #dir for labeled .tif
pred_dir <- 'pred_dir' #a place to predict to new data
lapply(FUN = function(x){dir.create(x)},
X = c(preproc_dir, umap_dir, lab_dir, pred_dir))
Tiling and Pre-processing
Define a 2-column matrix with coordinates corresponding to centroids at which to generate tiles. Specify tile size and a buffer to avoid edge effects during pre-processing neighborhood calculations. Specify the number of cores used during preprocessing. Note that parallel processing will only work on Unix systems for the native functions in paint2train at this time. Parallel processing in external functions, such as umap, will likely work on Windows systems and greatly speed up the dimension reduction step.
#some test coordinates
xcoords <- c(727495,
727919)
ycoords <- c(5175339,
5175408)
#bind into matrix
coord_mat <- cbind(xcoords, ycoords)
ls <- 30 #how big should the tiles be, this is the side length (in units of data, meters here)
buff <- 5 #buffer in units of data
pre_cores <- ifelse(.Platform$OS.type == 'unix', #how many cores to use for pre-processing
detectCores() - 1,
1)
umap_cores <- detectCores() - 1 #how many cores to use for UMAP dimension reduction
#make 30m tiles with 5m buffer (to avoid edge effects during pre-processing)
tile_at_coords(coords = coord_mat,
len_side = ls,
buffer = buff,
out_dir = preproc_dir,
img = image_dir,
ncores = pre_cores)
 Generate NDVI…
Generate NDVI…
mclapply(
FUN = ndvi_msavi,
X = list.files(preproc_dir, full.names = T),
mc.cores = pre_cores
)
 … and MSAVI values at tiles.
… and MSAVI values at tiles.
 Create edge detection layers by applying a Sobel filter across the first
three PCA axes of data generated thus far.
Create edge detection layers by applying a Sobel filter across the first
three PCA axes of data generated thus far.
mclapply(
FUN = sobel,
X = list.files(preproc_dir, full.names = T),
mc.cores = pre_cores
)
 Calculate mean and variance in 0.25, 0.5, and 1 meter neighborhoods for
first three PCA axes of data generated up to this point. The extents of
these neighborhoods are a critical tuning parameter that will need to be
paired with a particular dataset and modeling objective.
Calculate mean and variance in 0.25, 0.5, and 1 meter neighborhoods for
first three PCA axes of data generated up to this point. The extents of
these neighborhoods are a critical tuning parameter that will need to be
paired with a particular dataset and modeling objective.
neighborhoods <- c(0.25, 0.5, 1) #neighborhood radii in units of imagery
mclapply(
FUN = mean_var,
X = list.files(preproc_dir, full.names = T),
f_width = neighborhoods,
mc.cores = pre_cores
)
As a final pre-processing step, remove the buffers from around each tile.
mclapply(FUN = remove_buffer,
X = list.files(preproc_dir, full.names = T),
b = buff,
mc.cores = pre_cores)
Here is a sample of the outputs of the focal calculations described
above for the first tile.

Dimension Reduction
Next reduce the pre-processed layers into three dimension with the UMAP algorithm. This will facilitate identification of similar pixels in the painting app by conducting non-linear transformations of the data and mitigate the curse of dimensionality. For details on this method, please refer to the UWOT documentation.
lapply(FUN = umap_tile,
X = list.files(preproc_dir, full.names = TRUE),
out_dir = umap_dir,
n_threads = umap_cores,
n_sgd_threads = umap_cores)
Compare the original RGB tiles with outcomes from pre-processing and
dimension reduction. UMAP space is represented in similarity colors.

Running App Locally
Now we may label our data using the paint2train app. We first define the classes to label, assigning them an integer value, as well as a corresponding color palette to visualize labeled areas.
label_key <- list(Unknown = 0,
`Not woody` = 1,
`Woody` = 2)
pal <- c('royalblue',
'tan',
'green')
Provide these lists, location of the UMAP output, and label directory to the p2t function.
p2t(umap_dir = umap_dir,
label_dir = lab_dir,
label_key = label_key,
label_col = pal)
Data Labeling Process
Select imagery tiles from the dropdown menu found in the upper left.

Click on a region you wish to classify and adjust the Dissimilarity
Threshold to match the extent of the class to label.

Select which class to label from the Labeling Tools menu, then click
the Label painted areas button to save the painted pixels to that
class. After painting and labeling focal areas, fill the remaining
unlabeled points by click the FIll unlabeled as class button.

Adjust the color of painted areas from the Aesthetics Controls drop
down menu.

Manually edit pixels by using the draw tools (lower right). Draw a box
or polygon around the region you wish to edit, select the appropriate
class from the Select class to label menu, then click Label drawn
areas

Change the base imagery with the controls in the upper right.

Filter high and low value outlier pixels to brighten or darken base
imagery layers by adjusting the Baselayer quantiles in the
Aesthetics controls drop down menu.

Click and drag to move the controls as needed.

Model Training
Now generate a simple random forest model using the data labeled with p2t.
train_dat <- load_tdat(preproc_dir = preproc_dir,
label_dir = lab_dir,
ncores = pre_cores)
set.seed(123)
rf_mod <- ranger(label ~ .,
data = train_dat,
num.threads = umap_cores,
classification = TRUE)
print(rf_mod)
#Predict to some new data
og_ext <- extent(stack(image_dir))
mean_x <- mean(og_ext[1:2])
mean_y <- mean(og_ext[3:4])
tile_at_coords(coords = cbind(mean_x, mean_y - 25),
len_side = 25,
buffer = buff,
out_dir = pred_dir,
img = image_dir,
ncores = pre_cores)
pred_files <- list.files(pred_dir, full.names = TRUE)
pre_pipeline <- function(x, fs, b) {
ndvi_msavi(x)
sobel(x)
mean_var(x, f_width = fs)
remove_buffer(x, b)
}
mclapply(FUN = pre_pipeline,
X = pred_files,
mc.cores = pre_cores,
fs = neighborhoods,
b = buff)
pred_names <- colnames(train_dat[2:ncol(train_dat)])
new_dat <- do.call(rbind, mclapply(FUN =
function(x,p){setNames(as.data.frame(getValues(stack(x))), p)},
X = pred_files,
p = pred_names,
mc.cores = pre_cores))
preds <- predict(rf_mod, new_dat, num.threads = umap_cores)
pred_ras <- raster(pred_files[1])
values(pred_ras) <- preds$predictions
leaflet() %>%
addRasterRGB(stack(pred_files[1]),
r = 1, g = 2, b = 3,
group = 'RGB') %>%
addRasterImage(pred_ras,
project = FALSE,
opacity = 0.6,
colors = c('transparent','red'),
group = 'Canopy') %>%
addLayersControl(overlayGroups = c('Canopy'),
options = layersControlOptions(collapsed = FALSE))

Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
