README.md
In pwj6/tcmR: how to make herb to molecule
tcmR
tcmR是一个用于中药网络药理学数据挖掘和分析的R包。 其功能包括:解析天然药有效活性成分、天然药化合物作用靶点以及常见疾病靶点、天然药与疾病的共作用靶点。
tcmR is an R package for TCM network pharmacology data mining and analyzing. Its functions include: analyzing the effective active ingredients of natural medicines, the targets of natural medicine compounds, common disease targets, and the co-action targets of natural medicines and diseases.
Authors
[周晓北] (Zhou Xiaobei)
[彭文杰] (Peng Wenjie)
Installation
for Linux,Mac or Win user:
install.packages("devtools")
devtools::install_github("pwj6/tcmR")
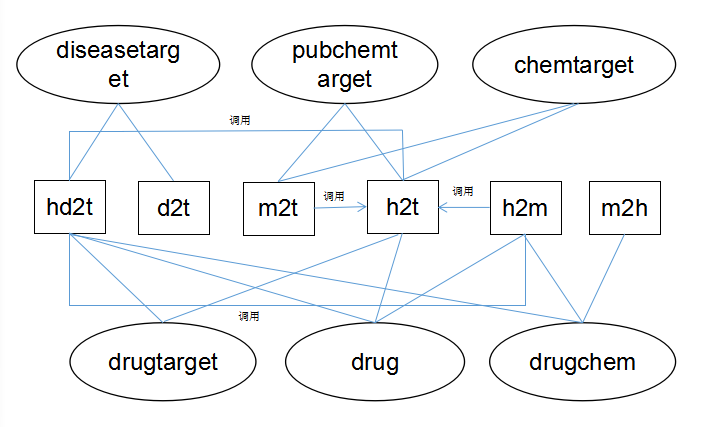
Relational graph of all functions and data
Usage
library(tcmR)
h2m(x='huangqin',type='pinyin')
"x" is the name of traditional Chinese medicine, which can be the Chinese name(type='chinese') or Chinese Pinyin of traditional Chinese medicine(type='pinyin'), or the Latin name of traditional Chinese medicine(type='latin').
The function of hd2t is to query the corresponding targets of diseases and traditional Chinese medicine, the compounds included in traditional Chinese medicine and the common targets of diseases and traditional Chinese medicine,including molecule,molecule_id,cid.The results are as follows:
$huangqin
molecule molecule_id cid
1: Cosmetin MOL000007 5280704
2: apigenin MOL000008 5280443
3: (+/-)-Isoborneol MOL000018 6321405
4: alpha-humulene MOL000024 5281520
5: beta-Selinene MOL000035 442393
---
139: 2,6,2',4'-tetrahydroxy-6'-methoxychaleone MOL002911 NA
140: darendoside B_qt MOL002924 NA
141: epiberberine MOL002897 NA
142: 3,7-dimethylnonane MOL012564 124556645
143: darendoside B MOL002923 NA
m2t(x=c('445858','31423'),type='cid',tSource="tcmsp")
"x" is the name of the compound, which can be the id(type='molecule_id'), English name (type='molecule')or cid of the compound(type='cid').
"tSource" is the source of compound targets,including tcmsp and pubchem.
The function of m2t is to transform compounds into their corresponding targets and related information.The results are as follows:
$`445858`
molecule molecule_id cid
1: FER MOL000360 445858
2: FER MOL000360 445858
3: FER MOL000360 445858
4: FER MOL000360 445858
5: FER MOL000360 445858
6: FER MOL000360 445858
7: FER MOL000360 445858
8: FER MOL000360 445858
9: FER MOL000360 445858
10: FER MOL000360 445858
11: FER MOL000360 445858
12: FER MOL000360 445858
13: FER MOL000360 445858
14: FER MOL000360 445858
15: FER MOL000360 445858
16: FER MOL000360 445858
fullname symbol
1: Alpha-2A adrenergic receptor ADRA2A
2: Chymotrypsinogen B CTRB1
3: Sodium-dependent noradrenaline transporter SLC6A2
4: Alpha-1A adrenergic receptor ADRA1A
5: Alpha-2B adrenergic receptor ADRA2B
6: Sodium-dependent dopamine transporter SLC6A3
7: Beta-2 adrenergic receptor ADRB2
8: Urokinase-type plasminogen activator PLAU
9: Heat shock protein HSP 90-alpha HSP90AA1
10: Leukotriene A-4 hydrolase LTA4H
11: Amine oxidase [flavin-containing] B MAOB
12: Amine oxidase [flavin-containing] A MAOA
13: Prostaglandin G/H synthase 1 PTGS1
14: cAMP-dependent protein kinase catalytic subunit alpha PRKACA
15: Prostaglandin G/H synthase 2 PTGS2
16: Nitric-oxide synthase, endothelial NOS3
$`31423`
molecule molecule_id cid fullname symbol
1: pyrene MOL005805 31423 Interleukin-8 <NA>
2: pyrene MOL005805 31423 UDP-glucuronosyltransferase 1-1 <NA>
3: pyrene MOL005805 31423 UDP-glucuronosyltransferase 1-6 <NA>
h2t(x=c('Ziziphi Spinosae Semen','Abri Herba'),type='latin',tSource="tcmsp",output='both')
"x" is exactly the same as x in h2m.
"tSource" is the source of traditional Chinese medicine targets,including tcmsp and pubchem.
"output" is how much information is output.output='symbol' includes molecule,molecule_id,cid,fullname and symbol;
output='both' includes herb,molecule,molecule_id,cid,fullname,symbol,ob and dl.
The function of h2t is to transform traditional Chinese medicine into its corresponding targets and related information.The results are as follows:
$`Ziziphi Spinosae Semen`
herb molecule molecule_id cid fullname symbol ob dl
1: Ziziphi Spinosae Semen Mairin MOL000211 64971 Progesterone receptor PGR 55.37707 0.776100
2: Ziziphi Spinosae Semen FER MOL000360 445858 Alpha-2A adrenergic receptor ADRA2A 39.55852 0.058069
3: Ziziphi Spinosae Semen FER MOL000360 445858 Chymotrypsinogen B CTRB1 39.55852 0.058069
4: Ziziphi Spinosae Semen FER MOL000360 445858 Sodium-dependent noradrenaline transporter SLC6A2 39.55852 0.058069
5: Ziziphi Spinosae Semen FER MOL000360 445858 Alpha-1A adrenergic receptor ADRA1A 39.55852 0.058069
---
144: Ziziphi Spinosae Semen zizyphusine MOL001546 NA Androgen receptor AR 41.52695 0.547170
145: Ziziphi Spinosae Semen zizyphusine MOL001546 NA Prostaglandin G/H synthase 1 PTGS1 41.52695 0.547170
146: Ziziphi Spinosae Semen zizyphusine MOL001546 NA Sodium channel protein type 5 subunit alpha SCN5A 41.52695 0.547170
147: Ziziphi Spinosae Semen zizyphusine MOL001546 NA Prostaglandin G/H synthase 2 PTGS2 41.52695 0.547170
148: Ziziphi Spinosae Semen alphitolic acid MOL001549 NA <NA> <NA> 23.38245 0.765810
$`Abri Herba`
herb molecule molecule_id cid fullname symbol ob dl
1: Abri Herba protocatechuic acid MOL000105 72 <NA> <NA> 25.366468 0.035092
2: Abri Herba protocatechuic acid MOL000105 72 Lysozyme E 25.366468 0.035092
3: Abri Herba protocatechuic acid MOL000105 72 Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase cobT 25.366468 0.035092
4: Abri Herba protocatechuic acid MOL000105 72 Maltase-glucoamylase, intestinal MGAM 25.366468 0.035092
5: Abri Herba protocatechuic acid MOL000105 72 Trypsin-3 PRSS3 25.366468 0.035092
---
365: Abri Herba kudzusapogenol A MOL003655 NA <NA> <NA> 16.848452 0.711260
366: Abri Herba Abrisapogenol F MOL013317 NA <NA> <NA> 16.366658 0.758840
367: Abri Herba Abrisapogenol B MOL013316 NA <NA> <NA> 14.743140 0.735340
368: Abri Herba Abrisapogenol E MOL003668 NA <NA> <NA> 17.578022 0.735780
369: Abri Herba Soyasaponin V MOL013328 NA <NA> <NA> 2.210538 0.048624
m2h(x=c('thalifendine','rosthorin A'),type='molecule',OB='10',DL='0.01')
"OB" and "DL" are the oral bioavailability and drug-like properties of compounds, respectively.Their default values are OB='30' and DL='0.18' respectively. You can also set thresholds according to your own needs.
The function of m2h is to query how many Chinese medicines a compound is contained in and the Latin names of these Chinese medicines.The results are as follows:
$thalifendine
herb molecule_id molecule cid ob dl
1: Phellodendri Amurensis Cortex MOL006422 thalifendine NA 44.41094 0.72588
2: Phellodendri Chinrnsis Cortex MOL006422 thalifendine NA 44.41094 0.72588
3: Thalictri Glandulosissimi Et MOL006422 thalifendine NA 44.41094 0.72588
4: Semiaquilegiae Radix MOL006422 thalifendine NA 44.41094 0.72588
$`rosthorin A`
herb molecule_id molecule cid ob dl
1: Ricini Semen MOL008144 rosthorin A NA 28.65893 0.56412
```r
d2t(x=c('C0009375','C0271979'),type='diseaseID',score=0.4)
"x" is the name of the disease, and can be the English name(type='disease') or id(type='diseaseID') of the disease.
"score" is the score of disease targets, which can be used to screen targets. The default value is 0.3, and you can also set thresholds according to your actual needs.
The function of d2t is to query the targets corresponding to the disease and relevant information.The results are as follows:
$C0009375
symbol score
1: APC 0.60
2: BCL2 0.56
3: CDKN1A 0.40
4: CTNNB1 0.40
5: DNMT1 0.52
6: EGFR 0.40
7: KRAS 0.40
8: MLH1 0.40
9: MMP9 0.54
10: MYC 0.53
11: PPARG 0.40
12: PTGS2 0.60
13: TP53 0.50
14: VEGFA 0.40
$C0271979
symbol score
1: HBA2 0.4
2: HBB 0.4
hd2t(hb=c('mahuang','huangqi'),Disease='C0019209',htype='pinyin',dtype='diseaseID',tSource='tcmsp')
"hb" is exactly the same as x in h2m;"Disease" is exactly the same as x in d2t;"htype" is exactly the same as type in h2m;"dtype" is exactly the same as type in d2t;"tSource" is exactly the same as tSource in h2t.
The function of hd2t is to query the corresponding targets of diseases and traditional Chinese medicine, the compounds included in traditional Chinese medicine and the common targets of diseases and traditional Chinese medicine.The results are as follows:
$mahuang
$mahuang$drug_target
herb molecule molecule_id cid fullname symbol ob dl
1: Ephedra Herba luteolin MOL000006 5280445 <NA> <NA> 36.16263 0.245520
2: Ephedra Herba luteolin MOL000006 5280445 Insulin receptor INSR 36.16263 0.245520
3: Ephedra Herba luteolin MOL000006 5280445 DNA topoisomerase 1 topA 36.16263 0.245520
4: Ephedra Herba luteolin MOL000006 5280445 Heme oxygenase 1 pbsA1 36.16263 0.245520
5: Ephedra Herba luteolin MOL000006 5280445 72 kDa type IV collagenase MMP2 36.16263 0.245520
---
3816: Ephedra Herba 1,5,8-trimethyl-1,2-dihydro-naphthalene MOL010789 137331948 Prostaglandin G/H synthase 1 PTGS1 53.22308 0.059796
3817: Ephedra Herba 1,5,8-trimethyl-1,2-dihydro-naphthalene MOL010789 137331948 D(1A) dopamine receptor DRD1 53.22308 0.059796
3818: Ephedra Herba 1,5,8-trimethyl-1,2-dihydro-naphthalene MOL010789 137331948 Prostaglandin G/H synthase 2 PTGS2 53.22308 0.059796
3819: Ephedra Herba 1,5,8-trimethyl-1,2-dihydro-naphthalene MOL010789 137331948 Nitric-oxide synthase, endothelial NOS3 53.22308 0.059796
3820: Ephedra Herba cinerolon MOL011873 92966642 <NA> <NA> 75.62580 0.036324
$mahuang$disease_target
symbol diseaseID disease score
1: A1BG C0019209 Hepatomegaly 0.3
2: AHR C0019209 Hepatomegaly 0.3
3: ALB C0019209 Hepatomegaly 0.3
4: ALDH1B1 C0019209 Hepatomegaly 0.3
5: CAMK2A C0019209 Hepatomegaly 0.3
6: CRYGD C0019209 Hepatomegaly 0.3
7: MAPK14 C0019209 Hepatomegaly 0.3
8: CYBA C0019209 Hepatomegaly 0.4
9: CYP1A1 C0019209 Hepatomegaly 0.3
10: CYP1A2 C0019209 Hepatomegaly 0.3
11: CYP1B1 C0019209 Hepatomegaly 0.3
12: ADAM3A C0019209 Hepatomegaly 0.3
13: DSCAM C0019209 Hepatomegaly 0.3
14: FGF12 C0019209 Hepatomegaly 0.3
15: NR5A2 C0019209 Hepatomegaly 0.3
16: HDC C0019209 Hepatomegaly 0.3
17: IGKC C0019209 Hepatomegaly 0.3
18: LEP C0019209 Hepatomegaly 0.3
19: LEPR C0019209 Hepatomegaly 0.3
20: MYO1B C0019209 Hepatomegaly 0.3
21: TRIM37 C0019209 Hepatomegaly 0.4
pwj6/tcmR documentation built on Dec. 22, 2021, 10:53 a.m.
tcmR
tcmR是一个用于中药网络药理学数据挖掘和分析的R包。 其功能包括:解析天然药有效活性成分、天然药化合物作用靶点以及常见疾病靶点、天然药与疾病的共作用靶点。
tcmR is an R package for TCM network pharmacology data mining and analyzing. Its functions include: analyzing the effective active ingredients of natural medicines, the targets of natural medicine compounds, common disease targets, and the co-action targets of natural medicines and diseases.
Authors
[周晓北] (Zhou Xiaobei)
[彭文杰] (Peng Wenjie)
Installation
for Linux,Mac or Win user:
install.packages("devtools")
devtools::install_github("pwj6/tcmR")
Relational graph of all functions and data

Usage
library(tcmR)
h2m(x='huangqin',type='pinyin')
"x" is the name of traditional Chinese medicine, which can be the Chinese name(type='chinese') or Chinese Pinyin of traditional Chinese medicine(type='pinyin'), or the Latin name of traditional Chinese medicine(type='latin').
The function of hd2t is to query the corresponding targets of diseases and traditional Chinese medicine, the compounds included in traditional Chinese medicine and the common targets of diseases and traditional Chinese medicine,including molecule,molecule_id,cid.The results are as follows:
$huangqin
molecule molecule_id cid
1: Cosmetin MOL000007 5280704
2: apigenin MOL000008 5280443
3: (+/-)-Isoborneol MOL000018 6321405
4: alpha-humulene MOL000024 5281520
5: beta-Selinene MOL000035 442393
---
139: 2,6,2',4'-tetrahydroxy-6'-methoxychaleone MOL002911 NA
140: darendoside B_qt MOL002924 NA
141: epiberberine MOL002897 NA
142: 3,7-dimethylnonane MOL012564 124556645
143: darendoside B MOL002923 NA
m2t(x=c('445858','31423'),type='cid',tSource="tcmsp")
"x" is the name of the compound, which can be the id(type='molecule_id'), English name (type='molecule')or cid of the compound(type='cid').
"tSource" is the source of compound targets,including tcmsp and pubchem.
The function of m2t is to transform compounds into their corresponding targets and related information.The results are as follows:
$`445858`
molecule molecule_id cid
1: FER MOL000360 445858
2: FER MOL000360 445858
3: FER MOL000360 445858
4: FER MOL000360 445858
5: FER MOL000360 445858
6: FER MOL000360 445858
7: FER MOL000360 445858
8: FER MOL000360 445858
9: FER MOL000360 445858
10: FER MOL000360 445858
11: FER MOL000360 445858
12: FER MOL000360 445858
13: FER MOL000360 445858
14: FER MOL000360 445858
15: FER MOL000360 445858
16: FER MOL000360 445858
fullname symbol
1: Alpha-2A adrenergic receptor ADRA2A
2: Chymotrypsinogen B CTRB1
3: Sodium-dependent noradrenaline transporter SLC6A2
4: Alpha-1A adrenergic receptor ADRA1A
5: Alpha-2B adrenergic receptor ADRA2B
6: Sodium-dependent dopamine transporter SLC6A3
7: Beta-2 adrenergic receptor ADRB2
8: Urokinase-type plasminogen activator PLAU
9: Heat shock protein HSP 90-alpha HSP90AA1
10: Leukotriene A-4 hydrolase LTA4H
11: Amine oxidase [flavin-containing] B MAOB
12: Amine oxidase [flavin-containing] A MAOA
13: Prostaglandin G/H synthase 1 PTGS1
14: cAMP-dependent protein kinase catalytic subunit alpha PRKACA
15: Prostaglandin G/H synthase 2 PTGS2
16: Nitric-oxide synthase, endothelial NOS3
$`31423`
molecule molecule_id cid fullname symbol
1: pyrene MOL005805 31423 Interleukin-8 <NA>
2: pyrene MOL005805 31423 UDP-glucuronosyltransferase 1-1 <NA>
3: pyrene MOL005805 31423 UDP-glucuronosyltransferase 1-6 <NA>
h2t(x=c('Ziziphi Spinosae Semen','Abri Herba'),type='latin',tSource="tcmsp",output='both')
"x" is exactly the same as x in h2m.
"tSource" is the source of traditional Chinese medicine targets,including tcmsp and pubchem.
"output" is how much information is output.output='symbol' includes molecule,molecule_id,cid,fullname and symbol;
output='both' includes herb,molecule,molecule_id,cid,fullname,symbol,ob and dl.
The function of h2t is to transform traditional Chinese medicine into its corresponding targets and related information.The results are as follows:
$`Ziziphi Spinosae Semen`
herb molecule molecule_id cid fullname symbol ob dl
1: Ziziphi Spinosae Semen Mairin MOL000211 64971 Progesterone receptor PGR 55.37707 0.776100
2: Ziziphi Spinosae Semen FER MOL000360 445858 Alpha-2A adrenergic receptor ADRA2A 39.55852 0.058069
3: Ziziphi Spinosae Semen FER MOL000360 445858 Chymotrypsinogen B CTRB1 39.55852 0.058069
4: Ziziphi Spinosae Semen FER MOL000360 445858 Sodium-dependent noradrenaline transporter SLC6A2 39.55852 0.058069
5: Ziziphi Spinosae Semen FER MOL000360 445858 Alpha-1A adrenergic receptor ADRA1A 39.55852 0.058069
---
144: Ziziphi Spinosae Semen zizyphusine MOL001546 NA Androgen receptor AR 41.52695 0.547170
145: Ziziphi Spinosae Semen zizyphusine MOL001546 NA Prostaglandin G/H synthase 1 PTGS1 41.52695 0.547170
146: Ziziphi Spinosae Semen zizyphusine MOL001546 NA Sodium channel protein type 5 subunit alpha SCN5A 41.52695 0.547170
147: Ziziphi Spinosae Semen zizyphusine MOL001546 NA Prostaglandin G/H synthase 2 PTGS2 41.52695 0.547170
148: Ziziphi Spinosae Semen alphitolic acid MOL001549 NA <NA> <NA> 23.38245 0.765810
$`Abri Herba`
herb molecule molecule_id cid fullname symbol ob dl
1: Abri Herba protocatechuic acid MOL000105 72 <NA> <NA> 25.366468 0.035092
2: Abri Herba protocatechuic acid MOL000105 72 Lysozyme E 25.366468 0.035092
3: Abri Herba protocatechuic acid MOL000105 72 Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase cobT 25.366468 0.035092
4: Abri Herba protocatechuic acid MOL000105 72 Maltase-glucoamylase, intestinal MGAM 25.366468 0.035092
5: Abri Herba protocatechuic acid MOL000105 72 Trypsin-3 PRSS3 25.366468 0.035092
---
365: Abri Herba kudzusapogenol A MOL003655 NA <NA> <NA> 16.848452 0.711260
366: Abri Herba Abrisapogenol F MOL013317 NA <NA> <NA> 16.366658 0.758840
367: Abri Herba Abrisapogenol B MOL013316 NA <NA> <NA> 14.743140 0.735340
368: Abri Herba Abrisapogenol E MOL003668 NA <NA> <NA> 17.578022 0.735780
369: Abri Herba Soyasaponin V MOL013328 NA <NA> <NA> 2.210538 0.048624
m2h(x=c('thalifendine','rosthorin A'),type='molecule',OB='10',DL='0.01')
"OB" and "DL" are the oral bioavailability and drug-like properties of compounds, respectively.Their default values are OB='30' and DL='0.18' respectively. You can also set thresholds according to your own needs.
The function of m2h is to query how many Chinese medicines a compound is contained in and the Latin names of these Chinese medicines.The results are as follows:
$thalifendine
herb molecule_id molecule cid ob dl
1: Phellodendri Amurensis Cortex MOL006422 thalifendine NA 44.41094 0.72588
2: Phellodendri Chinrnsis Cortex MOL006422 thalifendine NA 44.41094 0.72588
3: Thalictri Glandulosissimi Et MOL006422 thalifendine NA 44.41094 0.72588
4: Semiaquilegiae Radix MOL006422 thalifendine NA 44.41094 0.72588
$`rosthorin A`
herb molecule_id molecule cid ob dl
1: Ricini Semen MOL008144 rosthorin A NA 28.65893 0.56412
```r
d2t(x=c('C0009375','C0271979'),type='diseaseID',score=0.4)
"x" is the name of the disease, and can be the English name(type='disease') or id(type='diseaseID') of the disease.
"score" is the score of disease targets, which can be used to screen targets. The default value is 0.3, and you can also set thresholds according to your actual needs.
The function of d2t is to query the targets corresponding to the disease and relevant information.The results are as follows:
$C0009375
symbol score
1: APC 0.60
2: BCL2 0.56
3: CDKN1A 0.40
4: CTNNB1 0.40
5: DNMT1 0.52
6: EGFR 0.40
7: KRAS 0.40
8: MLH1 0.40
9: MMP9 0.54
10: MYC 0.53
11: PPARG 0.40
12: PTGS2 0.60
13: TP53 0.50
14: VEGFA 0.40
$C0271979
symbol score
1: HBA2 0.4
2: HBB 0.4
hd2t(hb=c('mahuang','huangqi'),Disease='C0019209',htype='pinyin',dtype='diseaseID',tSource='tcmsp')
"hb" is exactly the same as x in h2m;"Disease" is exactly the same as x in d2t;"htype" is exactly the same as type in h2m;"dtype" is exactly the same as type in d2t;"tSource" is exactly the same as tSource in h2t.
The function of hd2t is to query the corresponding targets of diseases and traditional Chinese medicine, the compounds included in traditional Chinese medicine and the common targets of diseases and traditional Chinese medicine.The results are as follows:
$mahuang
$mahuang$drug_target
herb molecule molecule_id cid fullname symbol ob dl
1: Ephedra Herba luteolin MOL000006 5280445 <NA> <NA> 36.16263 0.245520
2: Ephedra Herba luteolin MOL000006 5280445 Insulin receptor INSR 36.16263 0.245520
3: Ephedra Herba luteolin MOL000006 5280445 DNA topoisomerase 1 topA 36.16263 0.245520
4: Ephedra Herba luteolin MOL000006 5280445 Heme oxygenase 1 pbsA1 36.16263 0.245520
5: Ephedra Herba luteolin MOL000006 5280445 72 kDa type IV collagenase MMP2 36.16263 0.245520
---
3816: Ephedra Herba 1,5,8-trimethyl-1,2-dihydro-naphthalene MOL010789 137331948 Prostaglandin G/H synthase 1 PTGS1 53.22308 0.059796
3817: Ephedra Herba 1,5,8-trimethyl-1,2-dihydro-naphthalene MOL010789 137331948 D(1A) dopamine receptor DRD1 53.22308 0.059796
3818: Ephedra Herba 1,5,8-trimethyl-1,2-dihydro-naphthalene MOL010789 137331948 Prostaglandin G/H synthase 2 PTGS2 53.22308 0.059796
3819: Ephedra Herba 1,5,8-trimethyl-1,2-dihydro-naphthalene MOL010789 137331948 Nitric-oxide synthase, endothelial NOS3 53.22308 0.059796
3820: Ephedra Herba cinerolon MOL011873 92966642 <NA> <NA> 75.62580 0.036324
$mahuang$disease_target
symbol diseaseID disease score
1: A1BG C0019209 Hepatomegaly 0.3
2: AHR C0019209 Hepatomegaly 0.3
3: ALB C0019209 Hepatomegaly 0.3
4: ALDH1B1 C0019209 Hepatomegaly 0.3
5: CAMK2A C0019209 Hepatomegaly 0.3
6: CRYGD C0019209 Hepatomegaly 0.3
7: MAPK14 C0019209 Hepatomegaly 0.3
8: CYBA C0019209 Hepatomegaly 0.4
9: CYP1A1 C0019209 Hepatomegaly 0.3
10: CYP1A2 C0019209 Hepatomegaly 0.3
11: CYP1B1 C0019209 Hepatomegaly 0.3
12: ADAM3A C0019209 Hepatomegaly 0.3
13: DSCAM C0019209 Hepatomegaly 0.3
14: FGF12 C0019209 Hepatomegaly 0.3
15: NR5A2 C0019209 Hepatomegaly 0.3
16: HDC C0019209 Hepatomegaly 0.3
17: IGKC C0019209 Hepatomegaly 0.3
18: LEP C0019209 Hepatomegaly 0.3
19: LEPR C0019209 Hepatomegaly 0.3
20: MYO1B C0019209 Hepatomegaly 0.3
21: TRIM37 C0019209 Hepatomegaly 0.4
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
