README.md
In ryanburge/socsci: Some Basic Functions that I Use Frequently
SocSci: Functions for Analyzing Survey Data
Author
Ryan Burge https://www.ryanburge.net
Pkgdown site is available here: https://ryanburge.github.io/socsci/index.html
Instructor of Political Science, Eastern Illinois University, Charleston
IL
Installation
You can install:
-
the latest development version from GitHub with
r
install.packages("devtools")
devtools::install_github("ryanburge/socsci")
There are just a handful of functions to the package right now
Counting Things
I love the functionality of tabyl, but it doesn’t take a weight
variable. Here’s the simple version ct()
library(socsci)
cces <- read_csv("https://raw.githubusercontent.com/ryanburge/blocks/master/cces.csv")
cces %>%
ct(race)
#> # A tibble: 8 x 3
#> race n pct
#> <dbl> <int> <dbl>
#> 1 1 368 0.736
#> 2 2 54 0.108
#> 3 3 38 0.076
#> 4 4 13 0.026
#> 5 5 7 0.014
#> 6 6 9 0.018
#> 7 7 8 0.016
#> 8 8 3 0.006
Note that you are presented with a count column and a pct column.
Let’s add weights
cces %>%
ct(race, commonweight_vv)
#> # A tibble: 8 x 3
#> race n pct
#> <dbl> <dbl> <dbl>
#> 1 1 348. 0.758
#> 2 2 43.9 0.096
#> 3 3 34.0 0.074
#> 4 4 7.09 0.015
#> 5 5 3.87 0.008
#> 6 6 15.2 0.033
#> 7 7 6.33 0.014
#> 8 8 0.704 0.002
Notice that it’s pipeable. And if you don’t include the weight variable
then it won’t be calculated with a weight.
I’ve also added the ability to filter out the NA’s before the
calculation is made.
cces %>%
mutate(race2 = frcode(race == 1 ~ "White",
race == 2 ~ "Black",
race == 3 ~ "Hispanic",
race == 4 ~ "Asian")) %>%
ct(race2, commonweight_vv)
#> # A tibble: 5 x 3
#> race2 n pct
#> <fct> <dbl> <dbl>
#> 1 White 348. 0.758
#> 2 Black 43.9 0.096
#> 3 Hispanic 34.0 0.074
#> 4 Asian 7.09 0.015
#> 5 <NA> 26.1 0.057
cces %>%
mutate(race2 = frcode(race == 1 ~ "White",
race == 2 ~ "Black",
race == 3 ~ "Hispanic",
race == 4 ~ "Asian")) %>%
ct(race2, show_na = FALSE, commonweight_vv)
#> # A tibble: 4 x 3
#> race2 n pct
#> <fct> <dbl> <dbl>
#> 1 White 348. 0.804
#> 2 Black 43.9 0.101
#> 3 Hispanic 34.0 0.078
#> 4 Asian 7.09 0.016
This behavior is off by default, however.
Getting Confidence Intervals
Oftentimes in social science we like to see what our 95% confidence
intervals are, but that’s a lot of syntax. It’s easy with the mean_ci
function. I found the basic syntax on a Stack Overflow
post,
from the user
sboysel.
cces %>%
mean_ci(gender)
#> # A tibble: 1 x 7
#> mean sd n level se lower upper
#> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 1.54 0.499 500 0.05 0.0223 1.49 1.58
The default is a 95% confidence interval. However that can be changed
easily.
cces %>%
mean_ci(gender, ci = .84)
#> # A tibble: 1 x 7
#> mean sd n level se lower upper
#> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 1.54 0.499 500 0.16 0.0223 1.50 1.57
This can also take weights.
cces %>%
mean_ci(gender, ci = .84, wt = commonweight_vv)
#> # A tibble: 1 x 6
#> mean sd n se lower upper
#> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
#> 1 1.50 0.499 500 0.0223 1.47 1.53
Simple Mean and Median
I wanted a simple function to calculate the mean and the median. It
takes just one variable and computes both
statistics.
money1 <- read_csv("https://raw.githubusercontent.com/ryanburge/pls2003_sp17/master/sal_work.csv")
money1
#> # A tibble: 1,025 x 3
#> X1 salary names
#> <dbl> <dbl> <chr>
#> 1 1 14736 Darin Casem
#> 2 2 21261 Jaelyn Groesbeck
#> 3 3 16831 Theodis Butler
#> 4 4 34400 Joewid Rettig
#> 5 5 31239 Breianna Gilbert
#> 6 6 51580 Marcus Gray II
#> 7 7 49699 Berenice Garcia
#> 8 8 66805 Elijah Garrett
#> 9 9 49321 Jeremiah Bishop Jr
#> 10 10 67126 Sultana al-Jabbour
#> # ... with 1,015 more rows
money1 %>%
mean_med(salary)
#> # A tibble: 1 x 2
#> mean median
#> <dbl> <dbl>
#> 1 1247953. 35853
Two Value Correlations
Here’s a simple function that generates a pearson correlation of two
variables with a p-value.
x <- c(1, 2, 3, 7, 5, 777, 6, 411, 8)
y <- c(11, 23, 1, 4, 6, 22455, 34, 22, 22)
z <- c(34, 3, 21, 4555, 75, 2, 3334, 1122, 22312)
test <- data.frame(x,y,z) %>% as.tibble()
test %>%
filter(z > 10) %>%
corr(x,y)
#> # A tibble: 1 x 8
#> estimate statistic p.value n conf.low conf.high method alternative
#> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <chr> <chr>
#> 1 0.288 0.673 0.531 5 -0.594 0.856 Pearson's product-moment correlation two.sided
Bind Several Dataframes together
Oftentimes I make many little dataframes that I need to bind_rows to
put into one large dataframe. As long as those dataframes have the same
naming convention that can be done.
dd1 <- data.frame(a = 1, b = 2)
dd2 <- data.frame(a = 3, b = 4)
dd3 <- data.frame(a = 5, b = 6)
bind_df("dd")
#> a b
#> 1 1 2
#> 2 3 4
#> 3 5 6
Recode things and keep the factor levels
I recode all the time, but unfortunately when you recode from numeric to
character the factor levels are plotted in alphabetical order. There’s a
way around that now. This uses the case_when function from dplyr but
makes sure that the factors level are the same order of how they are
specified in the function.
I found this terrific
function
written by Dennis
YL, where he had the
same problem that I
had.
cces <- read_csv("https://raw.githubusercontent.com/ryanburge/cces/master/CCES%20for%20Methods/small_cces.csv")
graph <- cces %>%
mutate(pid_new = frcode(pid7 == 1 ~ "Strong Democrat",
pid7 == 2 ~ "Not Strong Democrat",
pid7 == 3 ~ "Lean Democrat",
pid7 == 4 ~ "Independent",
pid7 == 5 ~ "Lean Republican",
pid7 == 6 ~ "Not Strong Republican",
pid7 == 7 ~ "Strong Republican",
TRUE ~ "REMOVE")) %>%
ct(pid_new)
graph %>%
filter(pid_new != "REMOVE") %>%
ggplot(., aes(x = pid_new, y = pct)) +
geom_col()
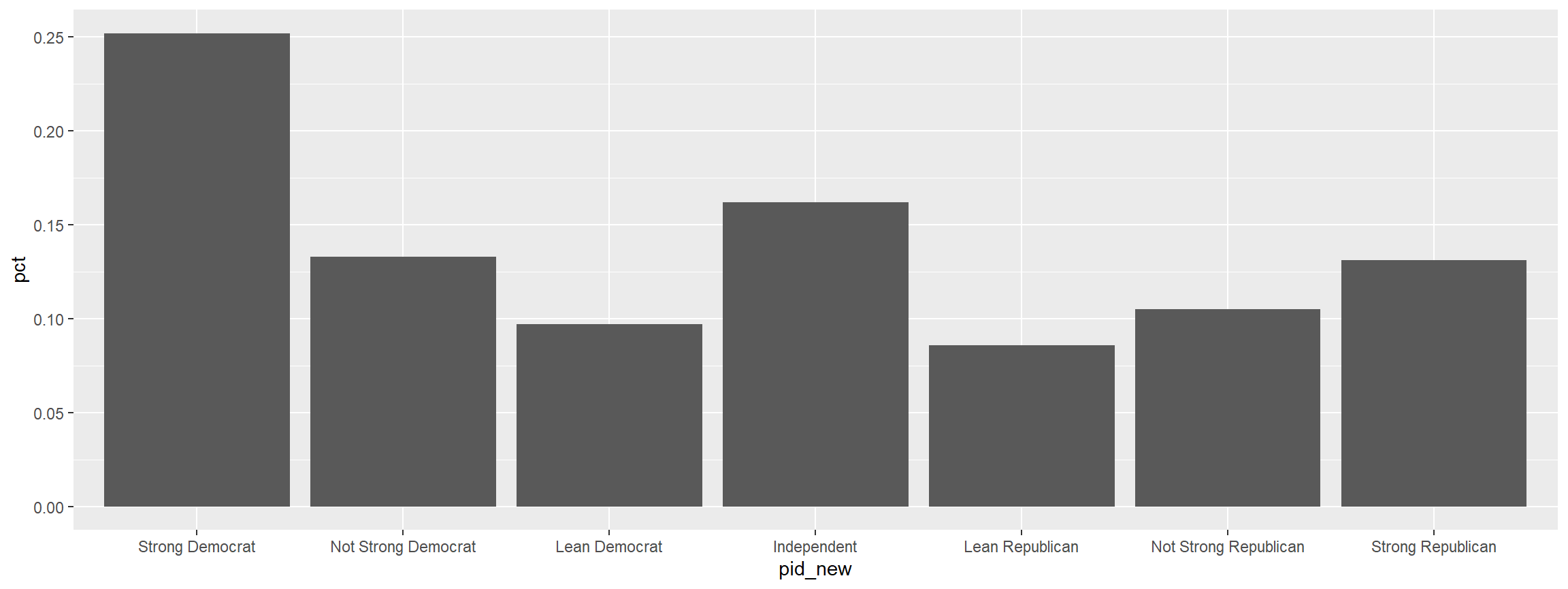
Making A Quick Crosstab Heatmap
Making a crosstab is one of the building blocks of social science
statistics. This function visualizes that crosstab. The first variable
is the one that is grouped and the second is the one that is counted
cces %>%
mutate(pid_new = frcode(pid7 == 1 ~ "Strong Democrat",
pid7 == 2 ~ "Not Strong Democrat",
pid7 == 3 ~ "Lean Democrat",
pid7 == 4 ~ "Independent",
pid7 == 5 ~ "Lean Republican",
pid7 == 6 ~ "Not Strong Republican",
pid7 == 7 ~ "Strong Republican",
TRUE ~ "All Others")) %>%
mutate(gender = frcode(gender ==1 ~ "Male",
gender ==2 ~ "Female")) %>%
xheat(gender, pid_new)
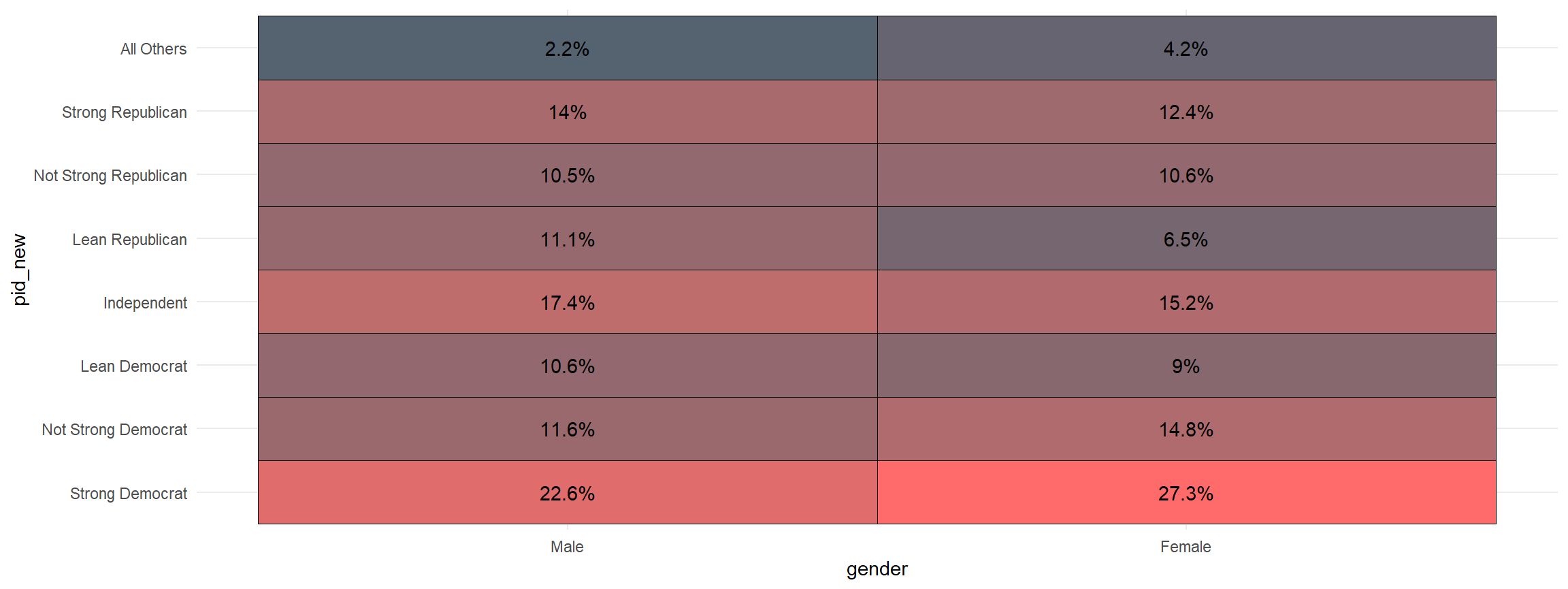
And, you can quickly add the sample size to the graph.
cces %>%
mutate(pid_new = frcode(pid7 == 1 ~ "Strong Democrat",
pid7 == 2 ~ "Not Strong Democrat",
pid7 == 3 ~ "Lean Democrat",
pid7 == 4 ~ "Independent",
pid7 == 5 ~ "Lean Republican",
pid7 == 6 ~ "Not Strong Republican",
pid7 == 7 ~ "Strong Republican",
TRUE ~ "All Others")) %>%
mutate(gender = frcode(gender ==1 ~ "Male",
gender ==2 ~ "Female")) %>%
xheat(gender, pid_new, count = TRUE)
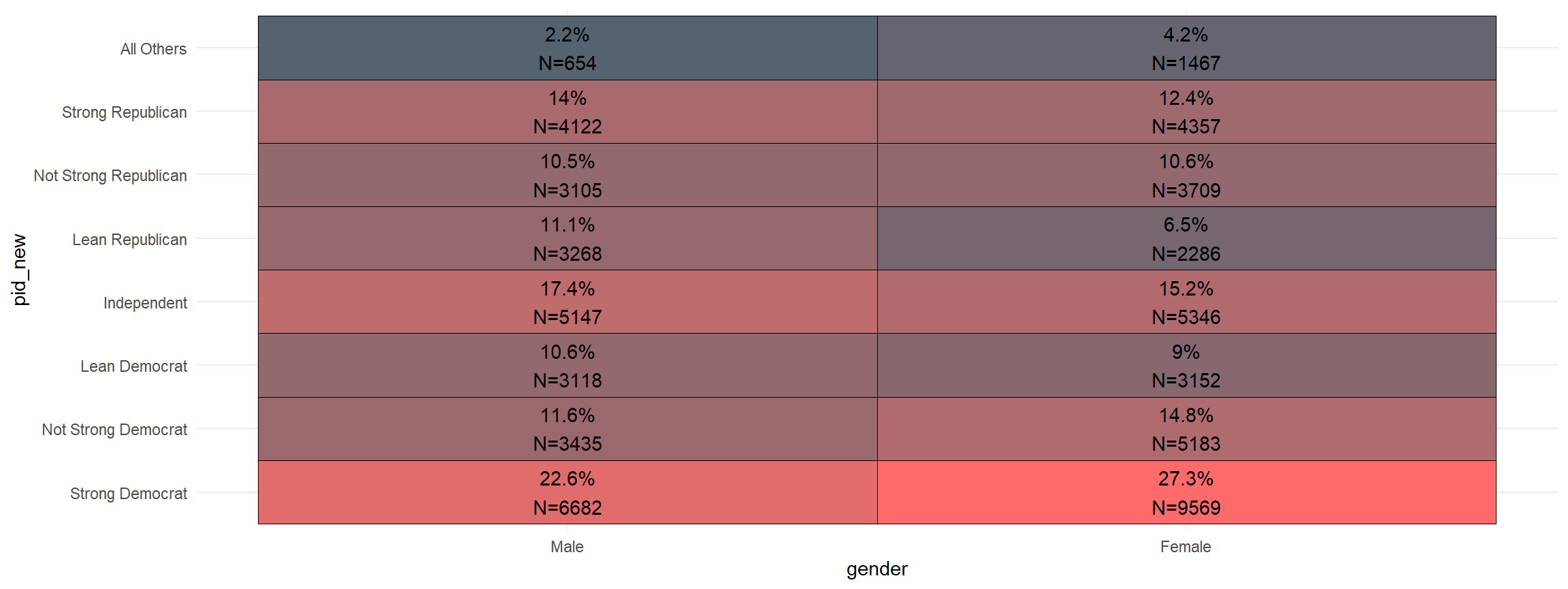
- let me know what you think on twitter
@ryanburge
ryanburge/socsci documentation built on June 6, 2020, 2:37 a.m.
SocSci: Functions for Analyzing Survey Data
Author
Ryan Burge https://www.ryanburge.net Pkgdown site is available here: https://ryanburge.github.io/socsci/index.html
Instructor of Political Science, Eastern Illinois University, Charleston IL
Installation
You can install:
-
the latest development version from GitHub with
r install.packages("devtools") devtools::install_github("ryanburge/socsci")
There are just a handful of functions to the package right now
Counting Things
I love the functionality of tabyl, but it doesn’t take a weight
variable. Here’s the simple version ct()
library(socsci)
cces <- read_csv("https://raw.githubusercontent.com/ryanburge/blocks/master/cces.csv")
cces %>%
ct(race)
#> # A tibble: 8 x 3
#> race n pct
#> <dbl> <int> <dbl>
#> 1 1 368 0.736
#> 2 2 54 0.108
#> 3 3 38 0.076
#> 4 4 13 0.026
#> 5 5 7 0.014
#> 6 6 9 0.018
#> 7 7 8 0.016
#> 8 8 3 0.006
Note that you are presented with a count column and a pct column.
Let’s add weights
cces %>%
ct(race, commonweight_vv)
#> # A tibble: 8 x 3
#> race n pct
#> <dbl> <dbl> <dbl>
#> 1 1 348. 0.758
#> 2 2 43.9 0.096
#> 3 3 34.0 0.074
#> 4 4 7.09 0.015
#> 5 5 3.87 0.008
#> 6 6 15.2 0.033
#> 7 7 6.33 0.014
#> 8 8 0.704 0.002
Notice that it’s pipeable. And if you don’t include the weight variable then it won’t be calculated with a weight.
I’ve also added the ability to filter out the NA’s before the calculation is made.
cces %>%
mutate(race2 = frcode(race == 1 ~ "White",
race == 2 ~ "Black",
race == 3 ~ "Hispanic",
race == 4 ~ "Asian")) %>%
ct(race2, commonweight_vv)
#> # A tibble: 5 x 3
#> race2 n pct
#> <fct> <dbl> <dbl>
#> 1 White 348. 0.758
#> 2 Black 43.9 0.096
#> 3 Hispanic 34.0 0.074
#> 4 Asian 7.09 0.015
#> 5 <NA> 26.1 0.057
cces %>%
mutate(race2 = frcode(race == 1 ~ "White",
race == 2 ~ "Black",
race == 3 ~ "Hispanic",
race == 4 ~ "Asian")) %>%
ct(race2, show_na = FALSE, commonweight_vv)
#> # A tibble: 4 x 3
#> race2 n pct
#> <fct> <dbl> <dbl>
#> 1 White 348. 0.804
#> 2 Black 43.9 0.101
#> 3 Hispanic 34.0 0.078
#> 4 Asian 7.09 0.016
This behavior is off by default, however.
Getting Confidence Intervals
Oftentimes in social science we like to see what our 95% confidence
intervals are, but that’s a lot of syntax. It’s easy with the mean_ci
function. I found the basic syntax on a Stack Overflow
post,
from the user
sboysel.
cces %>%
mean_ci(gender)
#> # A tibble: 1 x 7
#> mean sd n level se lower upper
#> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 1.54 0.499 500 0.05 0.0223 1.49 1.58
The default is a 95% confidence interval. However that can be changed easily.
cces %>%
mean_ci(gender, ci = .84)
#> # A tibble: 1 x 7
#> mean sd n level se lower upper
#> <dbl> <dbl> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 1.54 0.499 500 0.16 0.0223 1.50 1.57
This can also take weights.
cces %>%
mean_ci(gender, ci = .84, wt = commonweight_vv)
#> # A tibble: 1 x 6
#> mean sd n se lower upper
#> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
#> 1 1.50 0.499 500 0.0223 1.47 1.53
Simple Mean and Median
I wanted a simple function to calculate the mean and the median. It takes just one variable and computes both statistics.
money1 <- read_csv("https://raw.githubusercontent.com/ryanburge/pls2003_sp17/master/sal_work.csv")
money1
#> # A tibble: 1,025 x 3
#> X1 salary names
#> <dbl> <dbl> <chr>
#> 1 1 14736 Darin Casem
#> 2 2 21261 Jaelyn Groesbeck
#> 3 3 16831 Theodis Butler
#> 4 4 34400 Joewid Rettig
#> 5 5 31239 Breianna Gilbert
#> 6 6 51580 Marcus Gray II
#> 7 7 49699 Berenice Garcia
#> 8 8 66805 Elijah Garrett
#> 9 9 49321 Jeremiah Bishop Jr
#> 10 10 67126 Sultana al-Jabbour
#> # ... with 1,015 more rows
money1 %>%
mean_med(salary)
#> # A tibble: 1 x 2
#> mean median
#> <dbl> <dbl>
#> 1 1247953. 35853
Two Value Correlations
Here’s a simple function that generates a pearson correlation of two variables with a p-value.
x <- c(1, 2, 3, 7, 5, 777, 6, 411, 8)
y <- c(11, 23, 1, 4, 6, 22455, 34, 22, 22)
z <- c(34, 3, 21, 4555, 75, 2, 3334, 1122, 22312)
test <- data.frame(x,y,z) %>% as.tibble()
test %>%
filter(z > 10) %>%
corr(x,y)
#> # A tibble: 1 x 8
#> estimate statistic p.value n conf.low conf.high method alternative
#> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <chr> <chr>
#> 1 0.288 0.673 0.531 5 -0.594 0.856 Pearson's product-moment correlation two.sided
Bind Several Dataframes together
Oftentimes I make many little dataframes that I need to bind_rows to put into one large dataframe. As long as those dataframes have the same naming convention that can be done.
dd1 <- data.frame(a = 1, b = 2)
dd2 <- data.frame(a = 3, b = 4)
dd3 <- data.frame(a = 5, b = 6)
bind_df("dd")
#> a b
#> 1 1 2
#> 2 3 4
#> 3 5 6
Recode things and keep the factor levels
I recode all the time, but unfortunately when you recode from numeric to
character the factor levels are plotted in alphabetical order. There’s a
way around that now. This uses the case_when function from dplyr but
makes sure that the factors level are the same order of how they are
specified in the function.
I found this terrific function written by Dennis YL, where he had the same problem that I had.
cces <- read_csv("https://raw.githubusercontent.com/ryanburge/cces/master/CCES%20for%20Methods/small_cces.csv")
graph <- cces %>%
mutate(pid_new = frcode(pid7 == 1 ~ "Strong Democrat",
pid7 == 2 ~ "Not Strong Democrat",
pid7 == 3 ~ "Lean Democrat",
pid7 == 4 ~ "Independent",
pid7 == 5 ~ "Lean Republican",
pid7 == 6 ~ "Not Strong Republican",
pid7 == 7 ~ "Strong Republican",
TRUE ~ "REMOVE")) %>%
ct(pid_new)
graph %>%
filter(pid_new != "REMOVE") %>%
ggplot(., aes(x = pid_new, y = pct)) +
geom_col()

Making A Quick Crosstab Heatmap
Making a crosstab is one of the building blocks of social science statistics. This function visualizes that crosstab. The first variable is the one that is grouped and the second is the one that is counted
cces %>%
mutate(pid_new = frcode(pid7 == 1 ~ "Strong Democrat",
pid7 == 2 ~ "Not Strong Democrat",
pid7 == 3 ~ "Lean Democrat",
pid7 == 4 ~ "Independent",
pid7 == 5 ~ "Lean Republican",
pid7 == 6 ~ "Not Strong Republican",
pid7 == 7 ~ "Strong Republican",
TRUE ~ "All Others")) %>%
mutate(gender = frcode(gender ==1 ~ "Male",
gender ==2 ~ "Female")) %>%
xheat(gender, pid_new)

And, you can quickly add the sample size to the graph.
cces %>%
mutate(pid_new = frcode(pid7 == 1 ~ "Strong Democrat",
pid7 == 2 ~ "Not Strong Democrat",
pid7 == 3 ~ "Lean Democrat",
pid7 == 4 ~ "Independent",
pid7 == 5 ~ "Lean Republican",
pid7 == 6 ~ "Not Strong Republican",
pid7 == 7 ~ "Strong Republican",
TRUE ~ "All Others")) %>%
mutate(gender = frcode(gender ==1 ~ "Male",
gender ==2 ~ "Female")) %>%
xheat(gender, pid_new, count = TRUE)

- let me know what you think on twitter @ryanburge
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.



