README.md
In sciencepeak/rTCRBCRr: Repertoire Analysis of the Detected Clonotype
rTCRBCRr 
The goal of rTCRBCRr is to process the results from clonotyping tools
such as trust, mixcr, and immunoseq to analyze the clonotype repertoire
metrics
Installation
The package is accepted by the CRAN, you
can install the released version of rTCRBCRr from CRAN with:
install.packages("rTCRBCRr")
You can also install the development version from
GitHub with:
# install.packages("devtools")
devtools::install_github("sciencepeak/rTCRBCRr")
Example code
Attach packages
library("rTCRBCRr")
library("magrittr")
library("readr")
Read raw data files (trust generated for example) into a list of data frames
present_tool <- c("trust", "mixcr")[1]
example_data_directory <- system.file(paste("extdata", present_tool, sep = "/"), package = "rTCRBCRr")
input_paths <- dir(example_data_directory, full.names = TRUE)
input_files <- dir(example_data_directory, full.names = FALSE)
input_files
#> [1] "sample_01.tsv.bz2" "sample_02.tsv.bz2" "sample_03.tsv.bz2"
sample_names <- sub(".tsv.*", "", input_files)
sample_names
#> [1] "sample_01" "sample_02" "sample_03"
raw_clonotype_dataframe_list <- lapply(input_paths, readr::read_tsv) %>%
magrittr::set_names(., value = sample_names)
raw_clonotype_dataframe_list
#> $sample_01
#> # A tibble: 5,051 × 10
#> `#count` frequency CDR3nt CDR3aa V D J C cid cid_f…¹
#> <dbl> <dbl> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <dbl>
#> 1 1813 0.0664 TGTCAGAAGTAT… CQKYG… IGKV… . IGKJ… IGKC asse… 1
#> 2 1684 0.0833 TGCGCGAGAGAT… CARDA… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> 3 1648 0.0815 TGTGCGAGAGGG… CARGG… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> 4 1505 0.0551 TGTCAACAGCTT… CQQLS… IGKV… . IGKJ… IGKC asse… 1
#> 5 1081 0.0396 TGCATGCAAGGT… CMQGL… IGKV… . IGKJ… IGKC asse… 1
#> 6 895 0.0328 TGTCAACAGAGT… CQQSY… IGKV… . IGKJ… IGKC asse… 1
#> 7 866 0.0429 TGTGCGAGAGCC… CARAS… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> 8 811 0.0297 TGCATGCAAGCT… CMQAL… IGKV… . IGKJ… IGKC asse… 1
#> 9 767 0.0281 TGCTGCTCATAT… CCSYT… IGLV… . IGLJ… IGLC asse… 1
#> 10 761 0.0377 TGTGCTAGGGCG… CARAA… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> # … with 5,041 more rows, and abbreviated variable name ¹cid_full_length
#>
#> $sample_02
#> # A tibble: 5,915 × 10
#> `#count` frequency CDR3nt CDR3aa V D J C cid cid_f…¹
#> <dbl> <dbl> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <dbl>
#> 1 5392 0.0875 TGCATGCAAGCT… CMQAL… IGKV… . IGKJ… IGKC asse… 1
#> 2 4470 0.0725 TGCATGCAATCT… CMQSL… IGKV… . IGKJ… IGKC asse… 1
#> 3 3852 0.0625 TGTCAGCAGTAT… CQQYN… IGKV… . IGKJ… IGKC asse… 1
#> 4 2893 0.0470 TGCCAACGATAT… CQRYD… IGKV… . IGKJ… IGKC asse… 1
#> 5 2698 0.0438 TGTCAGCAGTAT… CQQYN… IGKV… . IGKJ… IGKC asse… 1
#> 6 1691 0.0509 TGTGCGAGACAA… CARQV… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> 7 1589 0.0478 TGTGCGAGACAC… CARHA… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> 8 1373 0.0414 TGTGCGAGACAG… CARQF… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> 9 1319 0.0397 TGTGCGGCAGAG… CAAEW… IGHV… IGHD… IGHJ… IGHG1 asse… 0
#> 10 1172 0.0353 TGTGCGAGGGGG… CARGV… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> # … with 5,905 more rows, and abbreviated variable name ¹cid_full_length
#>
#> $sample_03
#> # A tibble: 6,027 × 10
#> `#count` frequency CDR3nt CDR3aa V D J C cid cid_f…¹
#> <dbl> <dbl> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <dbl>
#> 1 14711 0.335 TGTACACTCCTG… CTLLV… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> 2 13909 0.341 TGCCAGTCCTAT… CQSYD… IGLV… . IGLJ… IGLC asse… 1
#> 3 6790 0.166 TGCAGCTCACAC… CSSHT… IGLV… . IGLJ… IGLC asse… 1
#> 4 4209 0.0959 TGTGCGAGAGCC… CARAL… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> 5 3727 0.0850 TGTGCGAGAGCC… CARAL… IGHV… IGHD… IGHJ… IGHA1 asse… 0
#> 6 910 0.0223 TGCAGCTCATAT… CSSYA… IGLV… . IGLJ… IGLC asse… 1
#> 7 771 0.0189 TGTGCAGCATGG… CAAWE… IGLV… . IGLJ… IGLC asse… 0
#> 8 768 0.0175 TGTGCGAGATTT… CARFF… IGHV… IGHD… IGHJ… IGHG3 asse… 1
#> 9 701 0.0172 TGCCAGTCCTAT… CQSYD… IGLV… . IGLJ… IGLC asse… 1
#> 10 628 0.0143 TGTGCGAGAGAT… CARDV… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> # … with 6,017 more rows, and abbreviated variable name ¹cid_full_length
Tidy up the clonotype dataframes
The tidy-up consists of four steps, namely four functions:
- format_clonotype_to_immunarch_style
- remove_nonproductive_CDR3aa
- annotate_chain_name_and_isotype_name
- merge_convergent_clonotype
# If you only want to test one sample, you can process the only sample as follows.
the_divergent_clonotype_dataframe <- raw_clonotype_dataframe_list[["sample_01"]] %>%
format_clonotype_to_immunarch_style(., clonotyping_tool = present_tool) %>%
remove_nonproductive_CDR3aa %>%
annotate_chain_name_and_isotype_name %>%
merge_convergent_clonotype
# Then the only one sample should be put into a list, element of which uses the sample name,
# because the later step need a named list of data frames as input.
divergent_clonotype_dataframe_list <- list(sample_01 = the_divergent_clonotype_dataframe)
# Otherwise, normally you will have multiple samples,
# then functional style of processing is preferred as follows.
divergent_clonotype_dataframe_list <- raw_clonotype_dataframe_list %>%
lapply(., format_clonotype_to_immunarch_style, clonotyping_tool = present_tool) %>%
lapply(., remove_nonproductive_CDR3aa) %>%
lapply(., annotate_chain_name_and_isotype_name) %>%
lapply(., merge_convergent_clonotype)
Calculate and merge repertoire metrics by chains for each sample in the list
This step consists of three functions.
# handle repertoire metrics for all the chains.
all_sample_all_chain_all_metrics_wide_format_dataframe_list <- the_divergent_clonotype_dataframe_list %>%
lapply(., compute_repertoire_metrics_by_chain_name)
all_sample_all_chain_all_metrics_wide_format_dataframe_list
#> $sample_01
#> diversity clonality richness evenness median
#> IGH 5.4161872 0.26283295 1552 0.7371670 0.0001509510
#> IGK 5.2677388 0.28931127 1656 0.7106887 0.0000908224
#> IGL 4.8949171 0.25490053 713 0.7450995 0.0004132231
#> TRA 4.3018929 0.06585582 100 0.9341442 0.0064377682
#> TRB 4.8358496 0.08467479 197 0.9153252 0.0036429872
#> TRD 0.6730117 0.02904941 2 0.9709506 0.5000000000
#> TRG 0.6931472 0.00000000 2 1.0000000 0.5000000000
#>
#> $sample_02
#> diversity clonality richness evenness median
#> IGH 5.3401961 0.28075859 1677 0.7192414 9.231905e-05
#> IGK 4.5087736 0.40269721 1898 0.5973028 4.043917e-05
#> IGL 4.7761543 0.28333416 784 0.7166658 1.851338e-04
#> TRA 3.7682140 0.07196974 58 0.9280303 1.398601e-02
#> TRB 3.8933793 0.11402336 81 0.8859766 9.174312e-03
#> TRD 0.5004024 0.27807191 2 0.7219281 5.000000e-01
#> TRG NA NA NA NA NA
#>
#> $sample_03
#> diversity clonality richness evenness median
#> IGH 3.3401373 0.5400036 1424 0.4599964 1.032684e-04
#> IGK 5.4716100 0.1767547 770 0.8232453 4.581552e-04
#> IGL 2.5747289 0.6218047 905 0.3781953 6.578731e-05
#> TRA 4.0981274 0.1194323 105 0.8805677 5.917160e-03
#> TRB 3.9120015 0.2282353 159 0.7717647 2.816901e-03
#> TRD NA NA NA NA NA
#> TRG 0.5982696 0.1368794 2 0.8631206 5.000000e-01
all_sample_all_chain_all_metrics_wide_format_dataframe <- all_sample_all_chain_all_metrics_wide_format_dataframe_list %>%
combine_all_sample_repertoire_metrics
all_sample_all_chain_all_metrics_wide_format_dataframe
#> sample_name item_name diversity clonality richness evenness median
#> 1 sample_01 IGH 5.4161872 0.26283295 1552 0.7371670 1.509510e-04
#> 2 sample_01 IGK 5.2677388 0.28931127 1656 0.7106887 9.082240e-05
#> 3 sample_01 IGL 4.8949171 0.25490053 713 0.7450995 4.132231e-04
#> 4 sample_01 TRA 4.3018929 0.06585582 100 0.9341442 6.437768e-03
#> 5 sample_01 TRB 4.8358496 0.08467479 197 0.9153252 3.642987e-03
#> 6 sample_01 TRD 0.6730117 0.02904941 2 0.9709506 5.000000e-01
#> 7 sample_01 TRG 0.6931472 0.00000000 2 1.0000000 5.000000e-01
#> 8 sample_02 IGH 5.3401961 0.28075859 1677 0.7192414 9.231905e-05
#> 9 sample_02 IGK 4.5087736 0.40269721 1898 0.5973028 4.043917e-05
#> 10 sample_02 IGL 4.7761543 0.28333416 784 0.7166658 1.851338e-04
#> 11 sample_02 TRA 3.7682140 0.07196974 58 0.9280303 1.398601e-02
#> 12 sample_02 TRB 3.8933793 0.11402336 81 0.8859766 9.174312e-03
#> 13 sample_02 TRD 0.5004024 0.27807191 2 0.7219281 5.000000e-01
#> 14 sample_02 TRG NA NA NA NA NA
#> 15 sample_03 IGH 3.3401373 0.54000361 1424 0.4599964 1.032684e-04
#> 16 sample_03 IGK 5.4716100 0.17675466 770 0.8232453 4.581552e-04
#> 17 sample_03 IGL 2.5747289 0.62180471 905 0.3781953 6.578731e-05
#> 18 sample_03 TRA 4.0981274 0.11943225 105 0.8805677 5.917160e-03
#> 19 sample_03 TRB 3.9120015 0.22823526 159 0.7717647 2.816901e-03
#> 20 sample_03 TRD NA NA NA NA NA
#> 21 sample_03 TRG 0.5982696 0.13687943 2 0.8631206 5.000000e-01
all_sample_all_chain_individual_metrics_dataframe_list <- all_sample_all_chain_all_metrics_wide_format_dataframe %>%
get_item_name_x_sample_name_for_each_metric
all_sample_all_chain_individual_metrics_dataframe_list
#> $diversity
#> sample_01 sample_02 sample_03
#> IGH 5.4161872 5.3401961 3.3401373
#> IGK 5.2677388 4.5087736 5.4716100
#> IGL 4.8949171 4.7761543 2.5747289
#> TRA 4.3018929 3.7682140 4.0981274
#> TRB 4.8358496 3.8933793 3.9120015
#> TRD 0.6730117 0.5004024 NA
#> TRG 0.6931472 NA 0.5982696
#>
#> $clonality
#> sample_01 sample_02 sample_03
#> IGH 0.26283295 0.28075859 0.5400036
#> IGK 0.28931127 0.40269721 0.1767547
#> IGL 0.25490053 0.28333416 0.6218047
#> TRA 0.06585582 0.07196974 0.1194323
#> TRB 0.08467479 0.11402336 0.2282353
#> TRD 0.02904941 0.27807191 NA
#> TRG 0.00000000 NA 0.1368794
#>
#> $richness
#> sample_01 sample_02 sample_03
#> IGH 1552 1677 1424
#> IGK 1656 1898 770
#> IGL 713 784 905
#> TRA 100 58 105
#> TRB 197 81 159
#> TRD 2 2 NA
#> TRG 2 NA 2
#>
#> $evenness
#> sample_01 sample_02 sample_03
#> IGH 0.7371670 0.7192414 0.4599964
#> IGK 0.7106887 0.5973028 0.8232453
#> IGL 0.7450995 0.7166658 0.3781953
#> TRA 0.9341442 0.9280303 0.8805677
#> TRB 0.9153252 0.8859766 0.7717647
#> TRD 0.9709506 0.7219281 NA
#> TRG 1.0000000 NA 0.8631206
#>
#> $median
#> sample_01 sample_02 sample_03
#> IGH 0.0001509510 9.231905e-05 1.032684e-04
#> IGK 0.0000908224 4.043917e-05 4.581552e-04
#> IGL 0.0004132231 1.851338e-04 6.578731e-05
#> TRA 0.0064377682 1.398601e-02 5.917160e-03
#> TRB 0.0036429872 9.174312e-03 2.816901e-03
#> TRD 0.5000000000 5.000000e-01 NA
#> TRG 0.5000000000 NA 5.000000e-01
Calculate and merge repertoire metrics by IGH isotypes for each sample in the list
This step consists of three functions.
# handle repertoire metrics all all the isotypes of IGH chain.
all_sample_IGH_chain_all_metrics_wide_format_dataframe_list <- the_divergent_clonotype_dataframe_list %>%
lapply(., calculate_IGH_isotype_proportion)
all_sample_IGH_chain_all_metrics_wide_format_dataframe_list
#> $sample_01
#> count proportion
#> IGHA 202 0.1828054299
#> IGHD 1 0.0009049774
#> IGHG 813 0.7357466063
#> IGHM 89 0.0805429864
#>
#> $sample_02
#> count proportion
#> IGHA 11 0.0080941869
#> IGHD 1 0.0007358352
#> IGHG 1074 0.7902869757
#> IGHM 273 0.2008830022
#>
#> $sample_03
#> count proportion
#> IGHA 416 0.343517754
#> IGHD 4 0.003303055
#> IGHG 775 0.639966969
#> IGHM 16 0.013212221
all_sample_IGH_chain_all_metrics_wide_format_dataframe <- all_sample_IGH_chain_all_metrics_wide_format_dataframe_list %>%
combine_all_sample_repertoire_metrics
all_sample_IGH_chain_all_metrics_wide_format_dataframe
#> sample_name item_name count proportion
#> 1 sample_01 IGHA 202 0.1828054299
#> 2 sample_01 IGHD 1 0.0009049774
#> 3 sample_01 IGHG 813 0.7357466063
#> 4 sample_01 IGHM 89 0.0805429864
#> 5 sample_02 IGHA 11 0.0080941869
#> 6 sample_02 IGHD 1 0.0007358352
#> 7 sample_02 IGHG 1074 0.7902869757
#> 8 sample_02 IGHM 273 0.2008830022
#> 9 sample_03 IGHA 416 0.3435177539
#> 10 sample_03 IGHD 4 0.0033030553
#> 11 sample_03 IGHG 775 0.6399669694
#> 12 sample_03 IGHM 16 0.0132122213
all_sample_IGH_chain_individual_metrics_dataframe_list <- all_sample_IGH_chain_all_metrics_wide_format_dataframe %>%
get_item_name_x_sample_name_for_each_metric
all_sample_IGH_chain_individual_metrics_dataframe_list
#> $count
#> sample_01 sample_02 sample_03
#> IGHA 202 11 416
#> IGHD 1 1 4
#> IGHG 813 1074 775
#> IGHM 89 273 16
#>
#> $proportion
#> sample_01 sample_02 sample_03
#> IGHA 0.1828054299 0.0080941869 0.343517754
#> IGHD 0.0009049774 0.0007358352 0.003303055
#> IGHG 0.7357466063 0.7902869757 0.639966969
#> IGHM 0.0805429864 0.2008830022 0.013212221
Clonotype repertoire metrics formulas
The repertoire metrics formula including richness, diversity (Shannon
entropy), evenness (Pielou’s eveness), clonality, and median (frequency
median) were defined as follows, where
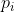 is the frequency
of
is the frequency
of
 in a sample with
in a sample with 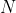 unique
clonotypes (Khunger, Rytlewski et
al. 2019,
Looney, Topacio-Hall et
al. 2020).
unique
clonotypes (Khunger, Rytlewski et
al. 2019,
Looney, Topacio-Hall et
al. 2020).
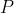 is the frequency vector
of unique clonotypes in a sample.
is the frequency vector
of unique clonotypes in a sample.





The function calculate_repertoire_metrics is essential to implement
the repertoire metrics formulas
calculate_repertoire_metrics
#> function (named_species_vector)
#> {
#> stopifnot(!duplicated(names(named_species_vector)))
#> species_vector <- named_species_vector
#> frequency_vector <- species_vector/sum(species_vector)
#> shannon_entropy <- -sum(frequency_vector * log(frequency_vector),
#> na.rm = TRUE)
#> richness_count <- length(species_vector)
#> pielou_evenness <- shannon_entropy/log(richness_count)
#> clonality_score <- 1 - pielou_evenness
#> frequency_median <- median(frequency_vector)
#> output_vector <- c(shannon_entropy, clonality_score, richness_count,
#> pielou_evenness, frequency_median)
#> names(output_vector) <- c("diversity", "clonality", "richness",
#> "evenness", "median")
#> output_vector
#> }
#> <bytecode: 0x000002071f9eb0f8>
#> <environment: namespace:rTCRBCRr>
Acknowledgements
The hexagon logo of
the package was created with the help of the package
hexSticker. The math
formula was written with the help of recognition tool
MyScript. The latex formula in markdown
was inspired by rmd4sci. The code
in this study was inspired by the UCSB R tutorial
note,
LymphoSeq script,
and vegan package.
sciencepeak/rTCRBCRr documentation built on March 11, 2023, 7:23 p.m.
rTCRBCRr 
The goal of rTCRBCRr is to process the results from clonotyping tools such as trust, mixcr, and immunoseq to analyze the clonotype repertoire metrics
Installation
The package is accepted by the CRAN, you can install the released version of rTCRBCRr from CRAN with:
install.packages("rTCRBCRr")
You can also install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("sciencepeak/rTCRBCRr")
Example code
Attach packages
library("rTCRBCRr")
library("magrittr")
library("readr")
Read raw data files (trust generated for example) into a list of data frames
present_tool <- c("trust", "mixcr")[1]
example_data_directory <- system.file(paste("extdata", present_tool, sep = "/"), package = "rTCRBCRr")
input_paths <- dir(example_data_directory, full.names = TRUE)
input_files <- dir(example_data_directory, full.names = FALSE)
input_files
#> [1] "sample_01.tsv.bz2" "sample_02.tsv.bz2" "sample_03.tsv.bz2"
sample_names <- sub(".tsv.*", "", input_files)
sample_names
#> [1] "sample_01" "sample_02" "sample_03"
raw_clonotype_dataframe_list <- lapply(input_paths, readr::read_tsv) %>%
magrittr::set_names(., value = sample_names)
raw_clonotype_dataframe_list
#> $sample_01
#> # A tibble: 5,051 × 10
#> `#count` frequency CDR3nt CDR3aa V D J C cid cid_f…¹
#> <dbl> <dbl> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <dbl>
#> 1 1813 0.0664 TGTCAGAAGTAT… CQKYG… IGKV… . IGKJ… IGKC asse… 1
#> 2 1684 0.0833 TGCGCGAGAGAT… CARDA… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> 3 1648 0.0815 TGTGCGAGAGGG… CARGG… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> 4 1505 0.0551 TGTCAACAGCTT… CQQLS… IGKV… . IGKJ… IGKC asse… 1
#> 5 1081 0.0396 TGCATGCAAGGT… CMQGL… IGKV… . IGKJ… IGKC asse… 1
#> 6 895 0.0328 TGTCAACAGAGT… CQQSY… IGKV… . IGKJ… IGKC asse… 1
#> 7 866 0.0429 TGTGCGAGAGCC… CARAS… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> 8 811 0.0297 TGCATGCAAGCT… CMQAL… IGKV… . IGKJ… IGKC asse… 1
#> 9 767 0.0281 TGCTGCTCATAT… CCSYT… IGLV… . IGLJ… IGLC asse… 1
#> 10 761 0.0377 TGTGCTAGGGCG… CARAA… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> # … with 5,041 more rows, and abbreviated variable name ¹cid_full_length
#>
#> $sample_02
#> # A tibble: 5,915 × 10
#> `#count` frequency CDR3nt CDR3aa V D J C cid cid_f…¹
#> <dbl> <dbl> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <dbl>
#> 1 5392 0.0875 TGCATGCAAGCT… CMQAL… IGKV… . IGKJ… IGKC asse… 1
#> 2 4470 0.0725 TGCATGCAATCT… CMQSL… IGKV… . IGKJ… IGKC asse… 1
#> 3 3852 0.0625 TGTCAGCAGTAT… CQQYN… IGKV… . IGKJ… IGKC asse… 1
#> 4 2893 0.0470 TGCCAACGATAT… CQRYD… IGKV… . IGKJ… IGKC asse… 1
#> 5 2698 0.0438 TGTCAGCAGTAT… CQQYN… IGKV… . IGKJ… IGKC asse… 1
#> 6 1691 0.0509 TGTGCGAGACAA… CARQV… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> 7 1589 0.0478 TGTGCGAGACAC… CARHA… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> 8 1373 0.0414 TGTGCGAGACAG… CARQF… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> 9 1319 0.0397 TGTGCGGCAGAG… CAAEW… IGHV… IGHD… IGHJ… IGHG1 asse… 0
#> 10 1172 0.0353 TGTGCGAGGGGG… CARGV… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> # … with 5,905 more rows, and abbreviated variable name ¹cid_full_length
#>
#> $sample_03
#> # A tibble: 6,027 × 10
#> `#count` frequency CDR3nt CDR3aa V D J C cid cid_f…¹
#> <dbl> <dbl> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <dbl>
#> 1 14711 0.335 TGTACACTCCTG… CTLLV… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> 2 13909 0.341 TGCCAGTCCTAT… CQSYD… IGLV… . IGLJ… IGLC asse… 1
#> 3 6790 0.166 TGCAGCTCACAC… CSSHT… IGLV… . IGLJ… IGLC asse… 1
#> 4 4209 0.0959 TGTGCGAGAGCC… CARAL… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> 5 3727 0.0850 TGTGCGAGAGCC… CARAL… IGHV… IGHD… IGHJ… IGHA1 asse… 0
#> 6 910 0.0223 TGCAGCTCATAT… CSSYA… IGLV… . IGLJ… IGLC asse… 1
#> 7 771 0.0189 TGTGCAGCATGG… CAAWE… IGLV… . IGLJ… IGLC asse… 0
#> 8 768 0.0175 TGTGCGAGATTT… CARFF… IGHV… IGHD… IGHJ… IGHG3 asse… 1
#> 9 701 0.0172 TGCCAGTCCTAT… CQSYD… IGLV… . IGLJ… IGLC asse… 1
#> 10 628 0.0143 TGTGCGAGAGAT… CARDV… IGHV… IGHD… IGHJ… IGHG1 asse… 1
#> # … with 6,017 more rows, and abbreviated variable name ¹cid_full_length
Tidy up the clonotype dataframes
The tidy-up consists of four steps, namely four functions:
- format_clonotype_to_immunarch_style
- remove_nonproductive_CDR3aa
- annotate_chain_name_and_isotype_name
- merge_convergent_clonotype
# If you only want to test one sample, you can process the only sample as follows.
the_divergent_clonotype_dataframe <- raw_clonotype_dataframe_list[["sample_01"]] %>%
format_clonotype_to_immunarch_style(., clonotyping_tool = present_tool) %>%
remove_nonproductive_CDR3aa %>%
annotate_chain_name_and_isotype_name %>%
merge_convergent_clonotype
# Then the only one sample should be put into a list, element of which uses the sample name,
# because the later step need a named list of data frames as input.
divergent_clonotype_dataframe_list <- list(sample_01 = the_divergent_clonotype_dataframe)
# Otherwise, normally you will have multiple samples,
# then functional style of processing is preferred as follows.
divergent_clonotype_dataframe_list <- raw_clonotype_dataframe_list %>%
lapply(., format_clonotype_to_immunarch_style, clonotyping_tool = present_tool) %>%
lapply(., remove_nonproductive_CDR3aa) %>%
lapply(., annotate_chain_name_and_isotype_name) %>%
lapply(., merge_convergent_clonotype)
Calculate and merge repertoire metrics by chains for each sample in the list
This step consists of three functions.
# handle repertoire metrics for all the chains.
all_sample_all_chain_all_metrics_wide_format_dataframe_list <- the_divergent_clonotype_dataframe_list %>%
lapply(., compute_repertoire_metrics_by_chain_name)
all_sample_all_chain_all_metrics_wide_format_dataframe_list
#> $sample_01
#> diversity clonality richness evenness median
#> IGH 5.4161872 0.26283295 1552 0.7371670 0.0001509510
#> IGK 5.2677388 0.28931127 1656 0.7106887 0.0000908224
#> IGL 4.8949171 0.25490053 713 0.7450995 0.0004132231
#> TRA 4.3018929 0.06585582 100 0.9341442 0.0064377682
#> TRB 4.8358496 0.08467479 197 0.9153252 0.0036429872
#> TRD 0.6730117 0.02904941 2 0.9709506 0.5000000000
#> TRG 0.6931472 0.00000000 2 1.0000000 0.5000000000
#>
#> $sample_02
#> diversity clonality richness evenness median
#> IGH 5.3401961 0.28075859 1677 0.7192414 9.231905e-05
#> IGK 4.5087736 0.40269721 1898 0.5973028 4.043917e-05
#> IGL 4.7761543 0.28333416 784 0.7166658 1.851338e-04
#> TRA 3.7682140 0.07196974 58 0.9280303 1.398601e-02
#> TRB 3.8933793 0.11402336 81 0.8859766 9.174312e-03
#> TRD 0.5004024 0.27807191 2 0.7219281 5.000000e-01
#> TRG NA NA NA NA NA
#>
#> $sample_03
#> diversity clonality richness evenness median
#> IGH 3.3401373 0.5400036 1424 0.4599964 1.032684e-04
#> IGK 5.4716100 0.1767547 770 0.8232453 4.581552e-04
#> IGL 2.5747289 0.6218047 905 0.3781953 6.578731e-05
#> TRA 4.0981274 0.1194323 105 0.8805677 5.917160e-03
#> TRB 3.9120015 0.2282353 159 0.7717647 2.816901e-03
#> TRD NA NA NA NA NA
#> TRG 0.5982696 0.1368794 2 0.8631206 5.000000e-01
all_sample_all_chain_all_metrics_wide_format_dataframe <- all_sample_all_chain_all_metrics_wide_format_dataframe_list %>%
combine_all_sample_repertoire_metrics
all_sample_all_chain_all_metrics_wide_format_dataframe
#> sample_name item_name diversity clonality richness evenness median
#> 1 sample_01 IGH 5.4161872 0.26283295 1552 0.7371670 1.509510e-04
#> 2 sample_01 IGK 5.2677388 0.28931127 1656 0.7106887 9.082240e-05
#> 3 sample_01 IGL 4.8949171 0.25490053 713 0.7450995 4.132231e-04
#> 4 sample_01 TRA 4.3018929 0.06585582 100 0.9341442 6.437768e-03
#> 5 sample_01 TRB 4.8358496 0.08467479 197 0.9153252 3.642987e-03
#> 6 sample_01 TRD 0.6730117 0.02904941 2 0.9709506 5.000000e-01
#> 7 sample_01 TRG 0.6931472 0.00000000 2 1.0000000 5.000000e-01
#> 8 sample_02 IGH 5.3401961 0.28075859 1677 0.7192414 9.231905e-05
#> 9 sample_02 IGK 4.5087736 0.40269721 1898 0.5973028 4.043917e-05
#> 10 sample_02 IGL 4.7761543 0.28333416 784 0.7166658 1.851338e-04
#> 11 sample_02 TRA 3.7682140 0.07196974 58 0.9280303 1.398601e-02
#> 12 sample_02 TRB 3.8933793 0.11402336 81 0.8859766 9.174312e-03
#> 13 sample_02 TRD 0.5004024 0.27807191 2 0.7219281 5.000000e-01
#> 14 sample_02 TRG NA NA NA NA NA
#> 15 sample_03 IGH 3.3401373 0.54000361 1424 0.4599964 1.032684e-04
#> 16 sample_03 IGK 5.4716100 0.17675466 770 0.8232453 4.581552e-04
#> 17 sample_03 IGL 2.5747289 0.62180471 905 0.3781953 6.578731e-05
#> 18 sample_03 TRA 4.0981274 0.11943225 105 0.8805677 5.917160e-03
#> 19 sample_03 TRB 3.9120015 0.22823526 159 0.7717647 2.816901e-03
#> 20 sample_03 TRD NA NA NA NA NA
#> 21 sample_03 TRG 0.5982696 0.13687943 2 0.8631206 5.000000e-01
all_sample_all_chain_individual_metrics_dataframe_list <- all_sample_all_chain_all_metrics_wide_format_dataframe %>%
get_item_name_x_sample_name_for_each_metric
all_sample_all_chain_individual_metrics_dataframe_list
#> $diversity
#> sample_01 sample_02 sample_03
#> IGH 5.4161872 5.3401961 3.3401373
#> IGK 5.2677388 4.5087736 5.4716100
#> IGL 4.8949171 4.7761543 2.5747289
#> TRA 4.3018929 3.7682140 4.0981274
#> TRB 4.8358496 3.8933793 3.9120015
#> TRD 0.6730117 0.5004024 NA
#> TRG 0.6931472 NA 0.5982696
#>
#> $clonality
#> sample_01 sample_02 sample_03
#> IGH 0.26283295 0.28075859 0.5400036
#> IGK 0.28931127 0.40269721 0.1767547
#> IGL 0.25490053 0.28333416 0.6218047
#> TRA 0.06585582 0.07196974 0.1194323
#> TRB 0.08467479 0.11402336 0.2282353
#> TRD 0.02904941 0.27807191 NA
#> TRG 0.00000000 NA 0.1368794
#>
#> $richness
#> sample_01 sample_02 sample_03
#> IGH 1552 1677 1424
#> IGK 1656 1898 770
#> IGL 713 784 905
#> TRA 100 58 105
#> TRB 197 81 159
#> TRD 2 2 NA
#> TRG 2 NA 2
#>
#> $evenness
#> sample_01 sample_02 sample_03
#> IGH 0.7371670 0.7192414 0.4599964
#> IGK 0.7106887 0.5973028 0.8232453
#> IGL 0.7450995 0.7166658 0.3781953
#> TRA 0.9341442 0.9280303 0.8805677
#> TRB 0.9153252 0.8859766 0.7717647
#> TRD 0.9709506 0.7219281 NA
#> TRG 1.0000000 NA 0.8631206
#>
#> $median
#> sample_01 sample_02 sample_03
#> IGH 0.0001509510 9.231905e-05 1.032684e-04
#> IGK 0.0000908224 4.043917e-05 4.581552e-04
#> IGL 0.0004132231 1.851338e-04 6.578731e-05
#> TRA 0.0064377682 1.398601e-02 5.917160e-03
#> TRB 0.0036429872 9.174312e-03 2.816901e-03
#> TRD 0.5000000000 5.000000e-01 NA
#> TRG 0.5000000000 NA 5.000000e-01
Calculate and merge repertoire metrics by IGH isotypes for each sample in the list
This step consists of three functions.
# handle repertoire metrics all all the isotypes of IGH chain.
all_sample_IGH_chain_all_metrics_wide_format_dataframe_list <- the_divergent_clonotype_dataframe_list %>%
lapply(., calculate_IGH_isotype_proportion)
all_sample_IGH_chain_all_metrics_wide_format_dataframe_list
#> $sample_01
#> count proportion
#> IGHA 202 0.1828054299
#> IGHD 1 0.0009049774
#> IGHG 813 0.7357466063
#> IGHM 89 0.0805429864
#>
#> $sample_02
#> count proportion
#> IGHA 11 0.0080941869
#> IGHD 1 0.0007358352
#> IGHG 1074 0.7902869757
#> IGHM 273 0.2008830022
#>
#> $sample_03
#> count proportion
#> IGHA 416 0.343517754
#> IGHD 4 0.003303055
#> IGHG 775 0.639966969
#> IGHM 16 0.013212221
all_sample_IGH_chain_all_metrics_wide_format_dataframe <- all_sample_IGH_chain_all_metrics_wide_format_dataframe_list %>%
combine_all_sample_repertoire_metrics
all_sample_IGH_chain_all_metrics_wide_format_dataframe
#> sample_name item_name count proportion
#> 1 sample_01 IGHA 202 0.1828054299
#> 2 sample_01 IGHD 1 0.0009049774
#> 3 sample_01 IGHG 813 0.7357466063
#> 4 sample_01 IGHM 89 0.0805429864
#> 5 sample_02 IGHA 11 0.0080941869
#> 6 sample_02 IGHD 1 0.0007358352
#> 7 sample_02 IGHG 1074 0.7902869757
#> 8 sample_02 IGHM 273 0.2008830022
#> 9 sample_03 IGHA 416 0.3435177539
#> 10 sample_03 IGHD 4 0.0033030553
#> 11 sample_03 IGHG 775 0.6399669694
#> 12 sample_03 IGHM 16 0.0132122213
all_sample_IGH_chain_individual_metrics_dataframe_list <- all_sample_IGH_chain_all_metrics_wide_format_dataframe %>%
get_item_name_x_sample_name_for_each_metric
all_sample_IGH_chain_individual_metrics_dataframe_list
#> $count
#> sample_01 sample_02 sample_03
#> IGHA 202 11 416
#> IGHD 1 1 4
#> IGHG 813 1074 775
#> IGHM 89 273 16
#>
#> $proportion
#> sample_01 sample_02 sample_03
#> IGHA 0.1828054299 0.0080941869 0.343517754
#> IGHD 0.0009049774 0.0007358352 0.003303055
#> IGHG 0.7357466063 0.7902869757 0.639966969
#> IGHM 0.0805429864 0.2008830022 0.013212221
Clonotype repertoire metrics formulas
The repertoire metrics formula including richness, diversity (Shannon
entropy), evenness (Pielou’s eveness), clonality, and median (frequency
median) were defined as follows, where
is the frequency
of
in a sample with
unique
clonotypes (Khunger, Rytlewski et
al. 2019,
Looney, Topacio-Hall et
al. 2020).
is the frequency vector
of unique clonotypes in a sample.
The function calculate_repertoire_metrics is essential to implement
the repertoire metrics formulas
calculate_repertoire_metrics
#> function (named_species_vector)
#> {
#> stopifnot(!duplicated(names(named_species_vector)))
#> species_vector <- named_species_vector
#> frequency_vector <- species_vector/sum(species_vector)
#> shannon_entropy <- -sum(frequency_vector * log(frequency_vector),
#> na.rm = TRUE)
#> richness_count <- length(species_vector)
#> pielou_evenness <- shannon_entropy/log(richness_count)
#> clonality_score <- 1 - pielou_evenness
#> frequency_median <- median(frequency_vector)
#> output_vector <- c(shannon_entropy, clonality_score, richness_count,
#> pielou_evenness, frequency_median)
#> names(output_vector) <- c("diversity", "clonality", "richness",
#> "evenness", "median")
#> output_vector
#> }
#> <bytecode: 0x000002071f9eb0f8>
#> <environment: namespace:rTCRBCRr>
Acknowledgements
The hexagon logo of the package was created with the help of the package hexSticker. The math formula was written with the help of recognition tool MyScript. The latex formula in markdown was inspired by rmd4sci. The code in this study was inspired by the UCSB R tutorial note, LymphoSeq script, and vegan package.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
