README.md
In stevehoang/pbayes: Convert p-values into Bayesian posterior probabilities
pbayes: Bayesian Probability Mapping for P-Values
pbayes is an R package designed to map p-values onto Bayesian posterior probabilities of the null hypothesis being false. It achieves this by modeling the distribution of p-values as a uniform-beta mixture. This approach allows users to interpret p-values from hypothesis tests within a Bayesian framework, offering a nuanced understanding of their results and enables many possibilities for meta-analysis.
Installation
You can install pbayes directly from GitHub using the remotes package:
# Install devtools if you haven't already
if (!requireNamespace("remotes", quietly = TRUE))
install.packages("remotes")
# Install pbayes from GitHub
remotes::install_github("stevehoang/pbayes")
Usage
This packages exposes several functions, but most users will only need to interface with pbayes. This function takes as input a collection of independent p-values (at least hundreds), and outputs 1) uniform-beta mixture distribution fitted to the data and 2) a posterior probability for each p-value that it corresponds to a non-uniform component of the fitted mixture. The latter value is interpreted as the posterior probability of the alternative hypothesis being true (equivalently, the posterior probability of the null hypothesis being false).
To demonstrate usage, we will simulate a distribution p-values and then use pbayes to fit a uniform-beta distribution to the data:
# Simulate p-values sampled from a mixture distribution -------------------
# set a random seed
set.seed(42)
# number of p-values to simulate
sample_size <- 10000
# set parameters
l0 <- 0.8 # mixing fraction of the uniform component of the mixture
l1 <- 0.2 # mixing fraction of the non-uniform beta component
r1 <- 0.5 # shape parameter 1 of the non-uniform beta component
s1 <- 3 # shape parameter 2 of the non-uniform beta component
# simulate p-values based on the parameters above
n_uniform <- sample_size * l0
n_nonuniform <- sample_size * l1
comp_uniform <- rbeta(n_uniform, 1, 1) # sample from a uniform distribution
comp_nonuniform <- rbeta(n_nonuniform, r1, s1) # sample from a non-uniform beta
p_sim <- c(comp_uniform, comp_nonuniform) # simulated p-values
hist(p_sim)
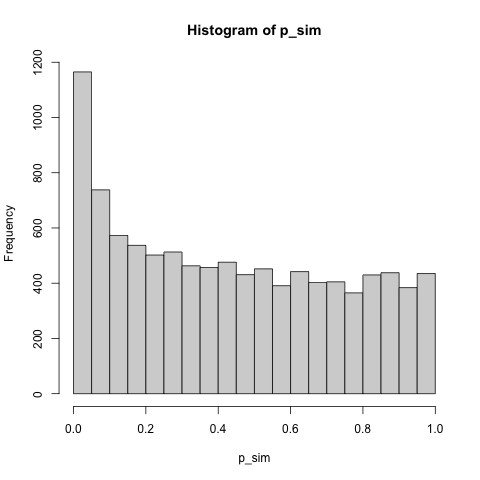
This type of p-value distribution is typical in biological "omic" experiments where hypothesis tests are performed across thousands of entities simultaneously (e.g., across genes).
Now we'll use pbayes to fit the mixure model and calculate posterior probabilities.
pb <- pbayes::pbayes(p_sim, n_cores = 5)
Let's look at the estimated parameters:
pb$mixture_model
l0 l1 r1 s1
0.8152414 0.1847586 0.5177831 3.4183191
attr(,"class")
[1] "betamix"
As expected, these parameter estimates are very close to the true parameters we specified above. We can also inspect a plot of the fitted components using the plot method of the pbayes object:
plot(pb)

Finally, we can inspect the relationship between the original p-values and the new posterior probabilities:
plot(pb$p_value, pb$posterior_prob)
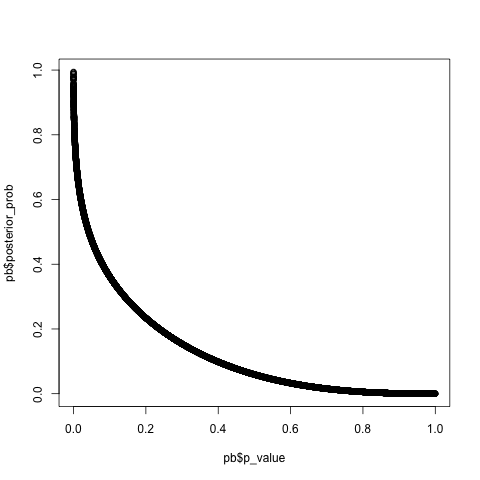
Small p-values map to large posterior probabilities. This is what we expect, since posterior probabilities near 1 indicate that the null hypothesis is likely false.
Additional information
The pbayes function offers many options for fitting, including controls on the computational resources, constraints on the number and type of beta components to use in the fit, different optimization algoritims, and more. Please see the function documentation for more information.
For more information on the theoretical basis of this pacakage, see the following:
Allison, D. B., et al. (2002). A mixture model approach for the analysis of microarray gene expression data. Computational Statistics & Data Analysis, 39(1), 1-20. https://doi.org/10.1016/S0167-9473(01)00046-9
Erikson S., et al. (2010). Composite hypothesis testing: and approach built on intersection-union tests and Bayesian posterior probabilities. In Guerra, R., and Goldstein, D. R., (Ed.), Meta-analysis and Combining Information in Genetics and Genomics. (pp. 83-93). Chapman & Hall/CRC.
stevehoang/pbayes documentation built on Aug. 21, 2024, 12:02 a.m.
pbayes: Bayesian Probability Mapping for P-Values
pbayes is an R package designed to map p-values onto Bayesian posterior probabilities of the null hypothesis being false. It achieves this by modeling the distribution of p-values as a uniform-beta mixture. This approach allows users to interpret p-values from hypothesis tests within a Bayesian framework, offering a nuanced understanding of their results and enables many possibilities for meta-analysis.
Installation
You can install pbayes directly from GitHub using the remotes package:
# Install devtools if you haven't already
if (!requireNamespace("remotes", quietly = TRUE))
install.packages("remotes")
# Install pbayes from GitHub
remotes::install_github("stevehoang/pbayes")
Usage
This packages exposes several functions, but most users will only need to interface with pbayes. This function takes as input a collection of independent p-values (at least hundreds), and outputs 1) uniform-beta mixture distribution fitted to the data and 2) a posterior probability for each p-value that it corresponds to a non-uniform component of the fitted mixture. The latter value is interpreted as the posterior probability of the alternative hypothesis being true (equivalently, the posterior probability of the null hypothesis being false).
To demonstrate usage, we will simulate a distribution p-values and then use pbayes to fit a uniform-beta distribution to the data:
# Simulate p-values sampled from a mixture distribution -------------------
# set a random seed
set.seed(42)
# number of p-values to simulate
sample_size <- 10000
# set parameters
l0 <- 0.8 # mixing fraction of the uniform component of the mixture
l1 <- 0.2 # mixing fraction of the non-uniform beta component
r1 <- 0.5 # shape parameter 1 of the non-uniform beta component
s1 <- 3 # shape parameter 2 of the non-uniform beta component
# simulate p-values based on the parameters above
n_uniform <- sample_size * l0
n_nonuniform <- sample_size * l1
comp_uniform <- rbeta(n_uniform, 1, 1) # sample from a uniform distribution
comp_nonuniform <- rbeta(n_nonuniform, r1, s1) # sample from a non-uniform beta
p_sim <- c(comp_uniform, comp_nonuniform) # simulated p-values
hist(p_sim)
This type of p-value distribution is typical in biological "omic" experiments where hypothesis tests are performed across thousands of entities simultaneously (e.g., across genes).
Now we'll use pbayes to fit the mixure model and calculate posterior probabilities.
pb <- pbayes::pbayes(p_sim, n_cores = 5)
Let's look at the estimated parameters:
pb$mixture_model
l0 l1 r1 s1
0.8152414 0.1847586 0.5177831 3.4183191
attr(,"class")
[1] "betamix"
As expected, these parameter estimates are very close to the true parameters we specified above. We can also inspect a plot of the fitted components using the plot method of the pbayes object:
plot(pb)
Finally, we can inspect the relationship between the original p-values and the new posterior probabilities:
plot(pb$p_value, pb$posterior_prob)
Small p-values map to large posterior probabilities. This is what we expect, since posterior probabilities near 1 indicate that the null hypothesis is likely false.
Additional information
The pbayes function offers many options for fitting, including controls on the computational resources, constraints on the number and type of beta components to use in the fit, different optimization algoritims, and more. Please see the function documentation for more information.
For more information on the theoretical basis of this pacakage, see the following:
Allison, D. B., et al. (2002). A mixture model approach for the analysis of microarray gene expression data. Computational Statistics & Data Analysis, 39(1), 1-20. https://doi.org/10.1016/S0167-9473(01)00046-9
Erikson S., et al. (2010). Composite hypothesis testing: and approach built on intersection-union tests and Bayesian posterior probabilities. In Guerra, R., and Goldstein, D. R., (Ed.), Meta-analysis and Combining Information in Genetics and Genomics. (pp. 83-93). Chapman & Hall/CRC.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
