README.md
In vertesy/MarkdownReportsDev: Generate Scientific Figures and Reports Easily
MarkdownReportsDev is discontinued.
Further development in MarkdownReports. See: #4.
MarkdownReportsDev was made when big changes were happening. The development reached its goals. Further development will be in MarkdownReports.
News
Version 4.1.0 is ready and installed by the default.
- See legacy/old version at: https://github.com/vertesy/MarkdownReports.v2.9.5
- See development version at: https://github.com/vertesy/MarkdownReportsDev
New features:
- Formatted session info Sessioninfo
- Updated dependencies
- Many functions are more stable and versatile
Old version is under MarkdownReports.LEGACY.VERSION.X.X.X.
What is MarkdownReports?
MarkdownReports is a set of R functions that allows you to generate precise figures easily, and create clean reports in markdown language about what you just discovered with your analysis script. It helps you to:
- Create scientifically accurate figures and save them automatically as vector graphic (.pdf), that you can use from presentation to posters anywhere.
- Note down your findings easily in a clear and nicely formatted way, parsed from your variables into english sentences.
- Link & display your figures automatically inside your report, right there where they are needed.
- Version your findings, annotating which parameters were used to reach certain results.
- Share your report with others via email, Github or a personal website.
Why did I make it & why you might like it too?
I do exploratory data analysis as a daily routine, and I have constant interaction with all sorts of people: supervisors, collaborators, colleagues, etc.
I often have to...
- Make figures quickly.
- ...write emails summarising the results (text & figures) of the last few days.
- ...find results from a couple of month back, with all tiny details (parameters used, etc).
- ...assemble each step I did that day into a logical story line, that others can understand at first glimpse, e.g.: I observed X; I controlled for Y; Hypothesised explanation A; Falsified it; Came up with explanation B; Tested & proven it...
For all of the above, my solution is MarkdownReports. I think its better than other solutions I found. Many of those like to combine source code with results, and many are too complex to use. Most of people I interact with are not interested in the source code, but are very keen on seeing my results from all possible angles and are asking detailed questions about the analysis.
Make figures quickly
- The philosophy of the package is to type little (but draw and save correctly annotated figures).
- Instead of specifying everything in lengthy commands (ala ggplot), plotting functions make use of sensible defaults (such as meaningful variable name, row names, column names, etc.)
- Both display and save each plot as
.pdf dynamically named (from variable names)
- Examples:
wboxplot() takes a list, used the variable name to set the filename and the title, list element names to set the x-axis labels, saves the file as variable name.pdf (or .png).
- All plot functions start with w, followed by the base plot name, such as
wplot(), wbarplot(), wpie(), wboxplot(),but also wvenn(), wvioplot_list(),wviostripchart_list().
- See more under: Discover 4 Yourself! (Below)
Write a report on the fly
Differences to Rmarkdown:
- It is intended for a different purpose:
- MarkdownReports is written for rapid progress reporting, whereas
- Rmarkdown is perfect for writing analytical explanations on "how do you analyse this?" and writing longer books.
- Much faster to report in MarkdownReports:
- You parse your report on the fly from directly your working script. (
- In Rmarkdown you would make a separate cleaned-up a script, that you then knit as a separate step.
- No hassle of executing computation in isolated code-blocks (and importing all relevant variables there). Your code is also a lot easier to follow because it is not split up in blocks.
Where does MarkdownReports stand out?
- Pure markdown output, compatible, simple and elegant layout.
- Integration of text, figures and tables with very few lines of extra code.
- Easy generation of precise figures (axis labels, coloring by filtering etc), a big enhancement over base graphics, while maintaining 90% of its syntax.
- Plots are both displayed and saved as a vector graphic (pdf), making it scalable for presentations, posters, etc
- Traceable results:
- PDF plots are labeled by the script generating them in the title field:
Filter.and.Stats by ExpressionAnalysis.R
- The report file is automatically named after the R-script, and date so that it is linked to the source code that generated it.
- Simply log all used settings into a markdown table by the
log_settings_MarkDown() and the md.LogSettingsFromList()functions.
- it natively exports tables from R to Markdown
- A timestamped subdirectory is created that you can backup once satisfied with your results.
- Github Compatibility:The generated report is easy to share on a GitHub wiki.
- It parses and writes full sentences to the report from operations you perform.
- For instance filter on gene expression level:
GeneExpression = rnorm(2000, mean = 100, sd=50);
MinExpression=125
PASS=filter_HP(GeneExpression, threshold = MinExpression)
and your report will have the summary: 30.7 % or 614 of 2000 entries in GeneExpression fall above a threshold value of: 125.
- Enhanced productivity features:
- Error bars are handled by
wbarplot() natively.
- Add an labels to bars in a barplot by
barplot_label().
- Native 2-D error bars in scatterplots
wplot() .
- Easy colour schemes by
wcolorize() from base, gplots and Rcolorbrewer.
- Add legends with the super short command
wlegend(colannot$categ), defining colors named after the categories of your data.
- It is autmatically created by
colannot = wcolorize(your.annotation, ReturnCategoriesToo = T), which you (can) anyways use to colour data points on, say, your scatterplot.
- Show filtering results with a one liner:
whist(rnorm(1000), vline = .5, filtercol = T).
- Although currently plotting is implemented as an enhanced base graphic, but the concept could easily be extended to ggplot.
Yet, you can still use ggplot, because you equally well save and report them by either
wplot_save_this() or the pdfA4plot_on() and pdfA4plot_off() functions.
- It is all achieved in ~ 1600 lines of well documented code compiled into a proper R-package.
Installation
Install directly from GitHub via devtools with one R command:
# install.packages("devtools"); # If you don't have it
require("devtools")
devtools::install_github(repo = "vertesy/MarkdownReportsDev")
...then simply load the package:
require("MarkdownReports")
Alternatively, you simply source it from the web.
This way function help will not work, and you will have no local copy of the code on your hard drive.
source("https://raw.githubusercontent.com/vertesy/MarkdownReportsDev/master/R/MarkdownReportsDev.R")
Discover 4 Yourself!
- Check out the wiki!
- See how easy it is to make customized figures.
- See the list of functions in the package.
- Check the example:
- Check out a dummy R script and the
- MarkDown report it generates inside this GitHub Repo.
- Browse the list of functions.
- Browse the code of the functions.
Learn about the markdown format
- See the power and simplicity of markdown format explained on Github
- Checkout some cool markdown editors, like MOU, Typora, or markdownpad.
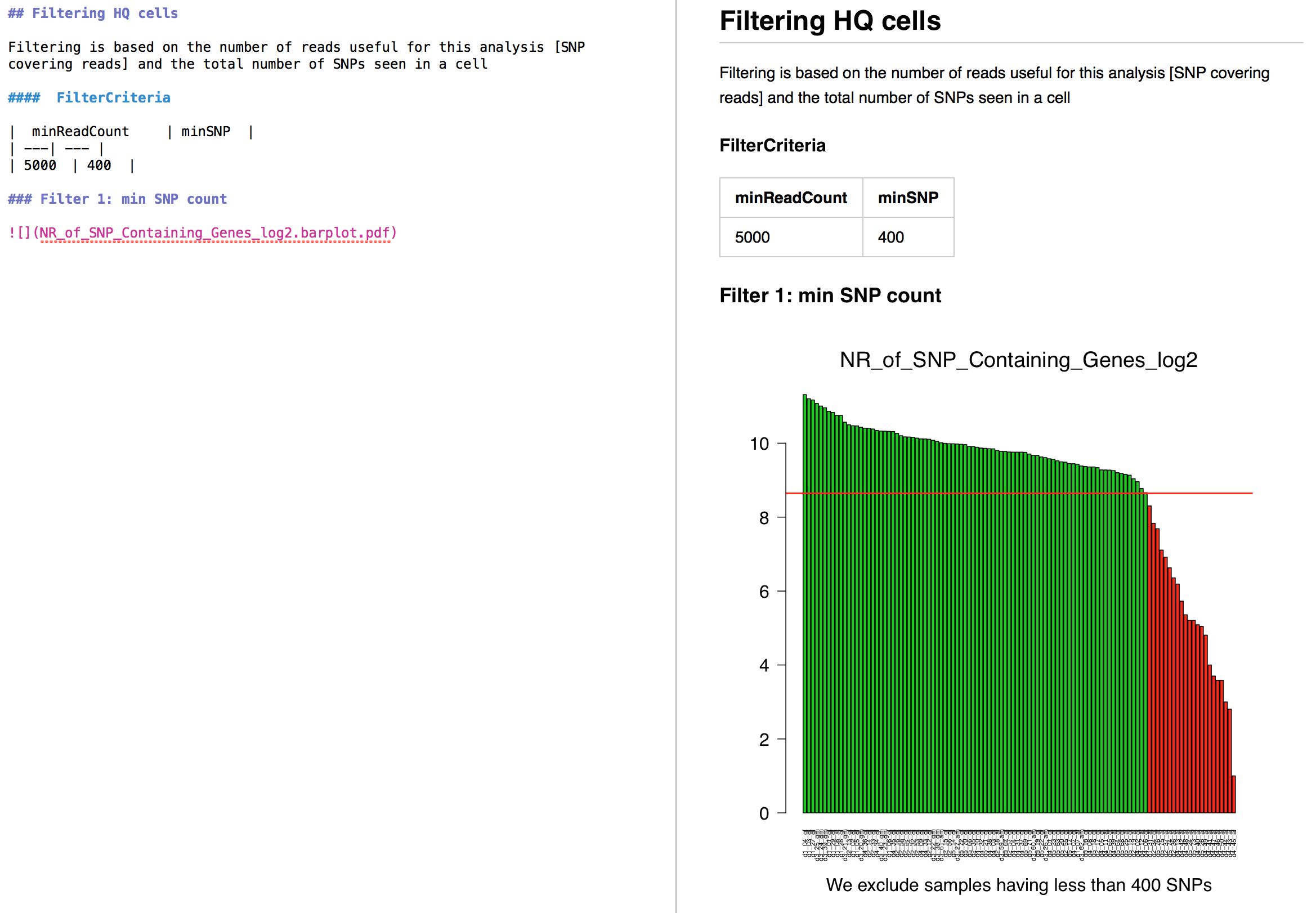
- See how these markdown reports are rendered:
Older News
[3.1.1 is under legacy now]
Version 3.1.1 is ready and installed by the default.
- See legacy/old version at: https://github.com/vertesy/MarkdownReports.v2.9.5
- See development version at: https://github.com/vertesy/MarkdownReportsDev
New features"
-
Function argument names now mirror the R base argument names (99%).
-
Think of xlb >>> xlab, or sub_ >>> sub
- This however breaks the compatibility with earlier versions, so you might need to replace some function arguments
-
The package now can also work with png images.
-
You can save files in png, which can be displayed inside the markdown file on windows 7.
-
You need to set b.usepng=T in setup_MarkdownReports: setup_MarkdownReports(OutDir = "/Users/...blabla....", b.usepng=T)
-
The package contains multiple other bug fixes:
-
Self consistency: some missing functions moved from CodeAndRoll.R
- Table writing functions
md.tableWriter.DF.w.dimnames() and md.tableWriter.VEC.w.names()
-
Enhancements:
-
filter_HP(), filter_LP(), filter_MidPass() show histogram
whist() can invite the above filter functions.- Numerous other small fixes.
Old version is under MarkdownReports.LEGACY.VERSION.
Cite it via its Digital Object Identifier (DOI):
Abel Vertesy. (2017, October 17). MarkdownReports: An R function library to create scientific figures and markdown reports easily. (Version v2.9.5). Zenodo. http://doi.org/10.5281/zenodo.594683
MarkdownReports is a project of @vertesy.
vertesy/MarkdownReportsDev documentation built on Nov. 15, 2021, 9:59 a.m.
MarkdownReportsDev is discontinued.
Further development in MarkdownReports. See: #4. MarkdownReportsDev was made when big changes were happening. The development reached its goals. Further development will be in MarkdownReports.
News
Version 4.1.0 is ready and installed by the default.
- See legacy/old version at: https://github.com/vertesy/MarkdownReports.v2.9.5
- See development version at: https://github.com/vertesy/MarkdownReportsDev
New features:
- Formatted session info Sessioninfo
- Updated dependencies
- Many functions are more stable and versatile
Old version is under MarkdownReports.LEGACY.VERSION.X.X.X.
What is MarkdownReports?
MarkdownReports is a set of R functions that allows you to generate precise figures easily, and create clean reports in markdown language about what you just discovered with your analysis script. It helps you to:
- Create scientifically accurate figures and save them automatically as vector graphic (.pdf), that you can use from presentation to posters anywhere.
- Note down your findings easily in a clear and nicely formatted way, parsed from your variables into english sentences.
- Link & display your figures automatically inside your report, right there where they are needed.
- Version your findings, annotating which parameters were used to reach certain results.
- Share your report with others via email, Github or a personal website.
Why did I make it & why you might like it too?
I do exploratory data analysis as a daily routine, and I have constant interaction with all sorts of people: supervisors, collaborators, colleagues, etc.
I often have to...
- Make figures quickly.
- ...write emails summarising the results (text & figures) of the last few days.
- ...find results from a couple of month back, with all tiny details (parameters used, etc).
- ...assemble each step I did that day into a logical story line, that others can understand at first glimpse, e.g.: I observed X; I controlled for Y; Hypothesised explanation A; Falsified it; Came up with explanation B; Tested & proven it...
For all of the above, my solution is MarkdownReports. I think its better than other solutions I found. Many of those like to combine source code with results, and many are too complex to use. Most of people I interact with are not interested in the source code, but are very keen on seeing my results from all possible angles and are asking detailed questions about the analysis.
Make figures quickly
- The philosophy of the package is to type little (but draw and save correctly annotated figures).
- Instead of specifying everything in lengthy commands (ala ggplot), plotting functions make use of sensible defaults (such as meaningful variable name, row names, column names, etc.)
- Both display and save each plot as
.pdfdynamically named (from variable names) - Examples:
wboxplot()takes a list, used thevariable nameto set the filename and the title,list element namesto set the x-axis labels, saves the file asvariable name.pdf(or.png). - All plot functions start with w, followed by the base plot name, such as
wplot(),wbarplot(),wpie(),wboxplot(),but alsowvenn(),wvioplot_list(),wviostripchart_list(). - See more under: Discover 4 Yourself! (Below)
Write a report on the fly
Differences to Rmarkdown:
- It is intended for a different purpose:
- MarkdownReports is written for rapid progress reporting, whereas
- Rmarkdown is perfect for writing analytical explanations on "how do you analyse this?" and writing longer books.
- Much faster to report in MarkdownReports:
- You parse your report on the fly from directly your working script. (
- In Rmarkdown you would make a separate cleaned-up a script, that you then knit as a separate step.
- No hassle of executing computation in isolated code-blocks (and importing all relevant variables there). Your code is also a lot easier to follow because it is not split up in blocks.
Where does MarkdownReports stand out?
- Pure markdown output, compatible, simple and elegant layout.
- Integration of text, figures and tables with very few lines of extra code.
- Easy generation of precise figures (axis labels, coloring by filtering etc), a big enhancement over base graphics, while maintaining 90% of its syntax.
- Plots are both displayed and saved as a vector graphic (pdf), making it scalable for presentations, posters, etc
- Traceable results:
- PDF plots are labeled by the script generating them in the title field:
Filter.and.Stats by ExpressionAnalysis.R - The report file is automatically named after the R-script, and date so that it is linked to the source code that generated it.
- Simply log all used settings into a markdown table by the
log_settings_MarkDown()and themd.LogSettingsFromList()functions. - it natively exports tables from R to Markdown
- A timestamped subdirectory is created that you can backup once satisfied with your results.
- Github Compatibility:The generated report is easy to share on a GitHub wiki.
- It parses and writes full sentences to the report from operations you perform.
- For instance filter on gene expression level:
GeneExpression = rnorm(2000, mean = 100, sd=50);
MinExpression=125
PASS=filter_HP(GeneExpression, threshold = MinExpression)
and your report will have the summary: 30.7 % or 614 of 2000 entries in GeneExpression fall above a threshold value of: 125.
- Enhanced productivity features:
- Error bars are handled by
wbarplot()natively. - Add an labels to bars in a barplot by
barplot_label(). - Native 2-D error bars in scatterplots
wplot(). - Easy colour schemes by
wcolorize()frombase,gplotsandRcolorbrewer. - Add legends with the super short command
wlegend(colannot$categ), defining colors named after the categories of your data.- It is autmatically created by
colannot = wcolorize(your.annotation, ReturnCategoriesToo = T), which you (can) anyways use to colour data points on, say, your scatterplot.
- It is autmatically created by
- Show filtering results with a one liner:
whist(rnorm(1000), vline = .5, filtercol = T). - Although currently plotting is implemented as an enhanced base graphic, but the concept could easily be extended to ggplot.
Yet, you can still use ggplot, because you equally well save and report them by either
wplot_save_this()or thepdfA4plot_on()andpdfA4plot_off()functions. - It is all achieved in ~ 1600 lines of well documented code compiled into a proper R-package.
Installation
Install directly from GitHub via devtools with one R command:
# install.packages("devtools"); # If you don't have it
require("devtools")
devtools::install_github(repo = "vertesy/MarkdownReportsDev")
...then simply load the package:
require("MarkdownReports")
Alternatively, you simply source it from the web. This way function help will not work, and you will have no local copy of the code on your hard drive.
source("https://raw.githubusercontent.com/vertesy/MarkdownReportsDev/master/R/MarkdownReportsDev.R")
Discover 4 Yourself!
- Check out the wiki!
- See how easy it is to make customized figures.
- See the list of functions in the package.
- Check the example:
- Check out a dummy R script and the
- MarkDown report it generates inside this GitHub Repo.
- Browse the list of functions.
- Browse the code of the functions.
Learn about the markdown format
- See the power and simplicity of markdown format explained on Github
- Checkout some cool markdown editors, like MOU, Typora, or markdownpad.
- See how these markdown reports are rendered:

Older News
[3.1.1 is under legacy now]
Version 3.1.1 is ready and installed by the default.
- See legacy/old version at: https://github.com/vertesy/MarkdownReports.v2.9.5
- See development version at: https://github.com/vertesy/MarkdownReportsDev
New features"
-
Function argument names now mirror the
R baseargument names (99%). -
Think of
xlb >>> xlab, orsub_ >>> sub - This however breaks the compatibility with earlier versions, so you might need to replace some function arguments
-
The package now can also work with png images.
-
You can save files in png, which can be displayed inside the markdown file on windows 7.
-
You need to set
b.usepng=Tinsetup_MarkdownReports:setup_MarkdownReports(OutDir = "/Users/...blabla....", b.usepng=T) -
The package contains multiple other bug fixes:
-
Self consistency: some missing functions moved from
CodeAndRoll.R - Table writing functions
md.tableWriter.DF.w.dimnames()andmd.tableWriter.VEC.w.names() -
Enhancements:
-
filter_HP(), filter_LP(), filter_MidPass()show histogram whist()can invite the above filter functions.- Numerous other small fixes.
Old version is under MarkdownReports.LEGACY.VERSION.
Cite it via its Digital Object Identifier (DOI):
Abel Vertesy. (2017, October 17). MarkdownReports: An R function library to create scientific figures and markdown reports easily. (Version v2.9.5). Zenodo. http://doi.org/10.5281/zenodo.594683
MarkdownReports is a project of @vertesy.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
