In vjcitn/BiocTNU: Interface to terra-notebook-utils
Introduction
The DataBiosphere project includes a vision
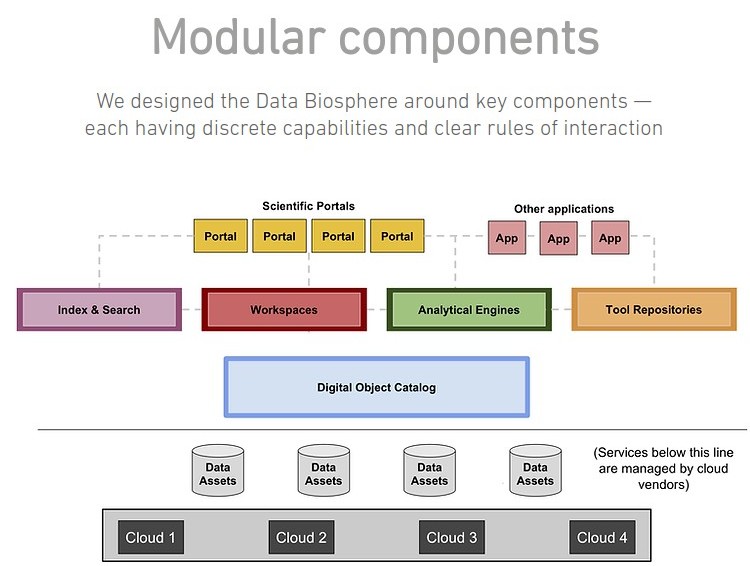
of which AnVIL/Terra forms a part.
The terra-notebook-utils[ python modules is described as a "Python
API and CLI providing utilities for working with DRS objects, VCF
files, and the Terra notebook environment."
This R package aims to provide a regulated interface between R and
terra-notebook-utils for use in AnVIL.
By "regulated" we mean that the entire python ecosystem used to work
with terra-notebook-utils is defined in a virtual environment. We
make some exceptions for the sake of demonstration, but, for example,
the drs_access command uses a very particular interface between R
and python, using the Bioconductor basilisk package.
Basic concepts
Installing in an AnVIL workspace, Oct 2022
As of 10/2022, AnVILTNU exists in a github repository. To install and
use properly with R in AnVIL
- use
BiocManager::install("vjcitn/AnVILTNU")
- verify that
library(AnVILTNU); example(drs_access) produces a
signed URL
Probing available features
Once installation has succeeded, we use basilisk-mediated commands
defined in the AnVILTNU package to probe or use terra-notebook-utils.
We can get the names of all modules available after importing
terra-notebook-utils.
library(AnVILTNU)
tnu_top()
We can also retrieve the help content for the python modules
subordinate to terra-notebook-utils.
tnu_help()
Generating signed URLS
The argument to drs_access is the google storage location of a CRAI
file.
uri <- "drs://dg.4503:dg.4503/17141a26-90e5-4160-9c20-89202b369431"
if (tnu_workspace_ok())
substr(drs_access(uri), 1, 80)
Copying DRS URLS
drs_copy is used to copy DRS URIs to a local file or to a google bucket. The
function requries a destination where the URLS should be copied to.
uris <- c(
"drs://dg.4503/15fdd543-9875-4edf-8bc2-22985473dab6",
"drs://dg.4503/3c861ec6-d810-4058-b851-c0b19dd5933e",
"drs://dg.4503/374a0ad9-b3a2-47f3-8860-5083b302e478"
)
if (tnu_workspace_ok())
drs_copy(uris, tempfile())
Generating information about DRS
There are a couple functions available that will provide information about the
DRS URLS provided. drs_head will query the DRS object and print the set number
of bytes while drs_info will return information about the DRS URIs.
if (tnu_workspace_ok()) {
drs_info(uris)
drs_head(uris, 100)
}
Firecloud functionality
Using the firecloud python module allows users to get information about tables
within their AnVIL workspace via the api. Below we demonstrate how a user can
utilize these functions via R / basilisk.
Table information
Within this package there are two function that display general table
information. tables() displays a tibble with 4 columns; table name,
row count, column count, and column names. table() requires a
specific table name and will display a tibble of the information
within that table.
if (tnu_workspace_ok()) {
tables()
table("test_table")
}
Additon or deletion of table information
A user may wish to upload a new table or add rows to a current table. The
function table_upload will allow the user to add information to a table within
their AnVIL workspace. We also provide deletion functionality which allows the
user the ability to delete single or multiple rows from a given table.
if (tnu_workspace_ok()) {
mycars <- head(mtcars) |>
dplyr::as_tibble(rownames = "model_id") |>
dplyr::mutate(model_id = gsub(" ", "_", model_id))
tables()
table("model")
table_upload(mycars)
table_delete_values("model", "Mazda_RX4")
table("model")
}
Shiny representation of tables
tables_gadget creates a shiny app that displays information about
the available tables in the workspace. A user could select a certain
table and on exit it will return that table within their R session as
a tibble.
if (tnu_workspace_ok())
tables_gadget()
Advanced use
The next section will demonstrate how to access other functions within
the firecloud module that may not be a part of this package. The
first thing to do is to get a basilisk session started. Then we will
import the module of interest, in this example it will be
firecloud. Using reticulate::py_help() displays the help page for
the available functions. Then you can just run the function of
interest using the specified arguments.
if (tnu_workspace_ok()) {
proc <- basiliskStart(bsklenv)
firecloud <- reticulate::import("firecloud")
reticulate::py_help(firecloud$api)
response <- firecloud$api$list_entity_types(tnu_workspace_namespace(),
tnu_workspace_name())
read.delim(text = response$text, sep = "\t", header = FALSE)
basiliskStop(proc)
}
We utilized rjsoncons::jmespath and jsonlite::fromJSON to make the
data more presentable within a tibble. This might be something of
interest to users who want to expand on other functions from the
modules used within this package.
Session information
sessionInfo()
vjcitn/BiocTNU documentation built on July 28, 2023, 8:09 p.m.
Introduction
The DataBiosphere project includes a vision

of which AnVIL/Terra forms a part.
The terra-notebook-utils[ python modules is described as a "Python API and CLI providing utilities for working with DRS objects, VCF files, and the Terra notebook environment."
This R package aims to provide a regulated interface between R and terra-notebook-utils for use in AnVIL.
By "regulated" we mean that the entire python ecosystem used to work
with terra-notebook-utils is defined in a virtual environment. We
make some exceptions for the sake of demonstration, but, for example,
the drs_access command uses a very particular interface between R
and python, using the Bioconductor basilisk package.
Basic concepts
Installing in an AnVIL workspace, Oct 2022
As of 10/2022, AnVILTNU exists in a github repository. To install and use properly with R in AnVIL
- use
BiocManager::install("vjcitn/AnVILTNU") - verify that
library(AnVILTNU); example(drs_access)produces a signed URL
Probing available features
Once installation has succeeded, we use basilisk-mediated commands defined in the AnVILTNU package to probe or use terra-notebook-utils. We can get the names of all modules available after importing terra-notebook-utils.
library(AnVILTNU) tnu_top()
We can also retrieve the help content for the python modules subordinate to terra-notebook-utils.
tnu_help()
Generating signed URLS
The argument to drs_access is the google storage location of a CRAI
file.
uri <- "drs://dg.4503:dg.4503/17141a26-90e5-4160-9c20-89202b369431" if (tnu_workspace_ok()) substr(drs_access(uri), 1, 80)
Copying DRS URLS
drs_copy is used to copy DRS URIs to a local file or to a google bucket. The
function requries a destination where the URLS should be copied to.
uris <- c( "drs://dg.4503/15fdd543-9875-4edf-8bc2-22985473dab6", "drs://dg.4503/3c861ec6-d810-4058-b851-c0b19dd5933e", "drs://dg.4503/374a0ad9-b3a2-47f3-8860-5083b302e478" ) if (tnu_workspace_ok()) drs_copy(uris, tempfile())
Generating information about DRS
There are a couple functions available that will provide information about the
DRS URLS provided. drs_head will query the DRS object and print the set number
of bytes while drs_info will return information about the DRS URIs.
if (tnu_workspace_ok()) { drs_info(uris) drs_head(uris, 100) }
Firecloud functionality
Using the firecloud python module allows users to get information about tables within their AnVIL workspace via the api. Below we demonstrate how a user can utilize these functions via R / basilisk.
Table information
Within this package there are two function that display general table
information. tables() displays a tibble with 4 columns; table name,
row count, column count, and column names. table() requires a
specific table name and will display a tibble of the information
within that table.
if (tnu_workspace_ok()) { tables() table("test_table") }
Additon or deletion of table information
A user may wish to upload a new table or add rows to a current table. The
function table_upload will allow the user to add information to a table within
their AnVIL workspace. We also provide deletion functionality which allows the
user the ability to delete single or multiple rows from a given table.
if (tnu_workspace_ok()) { mycars <- head(mtcars) |> dplyr::as_tibble(rownames = "model_id") |> dplyr::mutate(model_id = gsub(" ", "_", model_id)) tables() table("model") table_upload(mycars) table_delete_values("model", "Mazda_RX4") table("model") }
Shiny representation of tables
tables_gadget creates a shiny app that displays information about
the available tables in the workspace. A user could select a certain
table and on exit it will return that table within their R session as
a tibble.
if (tnu_workspace_ok()) tables_gadget()
Advanced use
The next section will demonstrate how to access other functions within
the firecloud module that may not be a part of this package. The
first thing to do is to get a basilisk session started. Then we will
import the module of interest, in this example it will be
firecloud. Using reticulate::py_help() displays the help page for
the available functions. Then you can just run the function of
interest using the specified arguments.
if (tnu_workspace_ok()) { proc <- basiliskStart(bsklenv) firecloud <- reticulate::import("firecloud") reticulate::py_help(firecloud$api) response <- firecloud$api$list_entity_types(tnu_workspace_namespace(), tnu_workspace_name()) read.delim(text = response$text, sep = "\t", header = FALSE) basiliskStop(proc) }
We utilized rjsoncons::jmespath and jsonlite::fromJSON to make the
data more presentable within a tibble. This might be something of
interest to users who want to expand on other functions from the
modules used within this package.
Session information
sessionInfo()
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
