README.md
In yanisb3812/DyrRegLog: Logistic Regression
DyrRegLog
Installing the package
To use the package, it has to be installed first.
devtools::install_github("yanisb3812/DyrRegLog")
Loading the library
When the package is installed, it can be loaded by using the library command from R.
library(DyrRegLog)
Dataset Import
A dataset is required to run the functions of this package. We'll be using the diabetes2 dataset.
data<-("diabetes2.csv")
Then we can use some commands from R to visualize our data.
str(data)
data.frame': 768 obs. of 9 variables:
$ Pregnancies : int 6 1 8 1 0 5 3 10 2 8 ...
$ Glucose : int 148 85 183 89 137 116 78 115 197 125 ...
$ BloodPressure : int 72 66 64 66 40 74 50 0 70 96 ...
$ SkinThickness : int 35 29 0 23 35 0 32 0 45 0 ...
$ Insulin : int 0 0 0 94 168 0 88 0 543 0 ...
$ BMI : num 33.6 26.6 23.3 28.1 43.1 25.6 31 35.3 30.5 0 ...
$ DiabetesPedigreeFunction: num 0.627 0.351 0.672 0.167 2.288 ...
$ Age : int 50 31 32 21 33 30 26 29 53 54 ...
$ Outcome : int 1 0 1 0 1 0 1 0 1 1 ...
Now we can detect our target variable and we see that all the variables are numeric.
It includes variables that can be the cause of the presence of diabetes, shown by the target variable "Outcome".
summary(data)
Pregnancies Glucose BloodPressure SkinThickness Insulin BMI DiabetesPedigreeFunction Age Outcome
Min. : 0.000 Min. : 0.0 Min. : 0.00 Min. : 0.00 Min. : 0.0 Min. : 0.00 Min. :0.0780 Min. :21.00 Min. :0.000
1st Qu.: 1.000 1st Qu.: 99.0 1st Qu.: 62.00 1st Qu.: 0.00 1st Qu.: 0.0 1st Qu.:27.30 1st Qu.:0.2437 1st Qu.:24.00 1st Qu.:0.000
Median : 3.000 Median :117.0 Median : 72.00 Median :23.00 Median : 30.5 Median :32.00 Median :0.3725 Median :29.00 Median :0.000
Mean : 3.845 Mean :120.9 Mean : 69.11 Mean :20.54 Mean : 79.8 Mean :31.99 Mean :0.4719 Mean :33.24 Mean :0.349
3rd Qu.: 6.000 3rd Qu.:140.2 3rd Qu.: 80.00 3rd Qu.:32.00 3rd Qu.:127.2 3rd Qu.:36.60 3rd Qu.:0.6262 3rd Qu.:41.00 3rd Qu.:1.000
Max. :17.000 Max. :199.0 Max. :122.00 Max. :99.00 Max. :846.0 Max. :67.10 Max. :2.4200 Max. :81.00 Max. :1.000
We can see that our variables are not on the same scale, we'll resolve that later on.
```
After that, we'll define a size of sample.
```
smp_size <- floor(0.70 * nrow(data))
```
Here, we selected 70% of the data size.
Then, we divide our sample into a training data and a test data randomly.
```
set.seed(29112021)
train_ind <- sample(seq_len(nrow(data)), size = smp_size)
data_train <- data[train_ind, ]
data_test <- data[-train_ind, ]
```
Now we can call the fit function of DyrRegLog package to fit our model to a Logistic Regression using the stochastic gradient descent algorithm.
test_fit = fit(formula = Outcome~. , data = data , eta = 0.3 , iter_Max = 10000 , mode = "Batch_Simple" , batch_size = 20, nb_Coeurs = 1 )
We can also visualize the plot of the deviances.
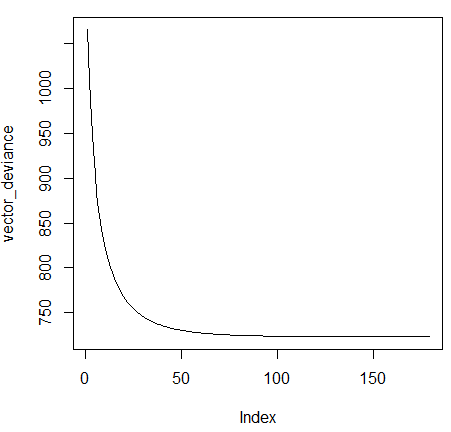
The print and summary functions are overridden.
So, if we print the variable containing the fit function, it will display the formula and the coefficients.
print(test_fit)
Formula : ~ Outcome .
Coefficients :
Pregnancies Glucose BloodPressure SkinThickness Insulin BMI DiabetesPedigreeFunction Age Intercept
1 0.4080121 1.110398 -0.2523509 0.005541574 -0.1286005 0.7017453 0.3108529 0.1803396 -0.8660777
It is the same thing for the summary function, but it will display the last value of
the deviance, the number of tierations and the time of execution of the algorithm.
summary(test_fit)
Last deviance value : 723.4693
Number of iterations : 180
Computing time : 0.35
Then we can predict our target variable with the predict function.
test_predict=predict(data_test[,-9], test_fit,sortie="Class")
print(test_predict)
[1] 0 1 0 0 1 0 1 1 1 1 1 1 1 1 1 1 1 1 0 1 1 0 1 1 0 1 0 1 0 1 1 1 0 1 0 0 1
[38] 1 0 1 0 1 0 1 0 1 1 0 0 1 1 1 1 1 0 1 1 1 1 0 1 1 1 1 1 0 1 1 0 1 1 1 1 1
[75] 1 1 0 0 1 1 0 0 1 1 1 1 1 0 1 1 1 0 1 1 1 1 1 0 1 1 1 1 1 1 1 1 1 1 0 1 1
[112] 1 1 1 1 1 1 0 0 1 0 1 1 1 0 1 1 1 1 1 0 1 1 0 0 1 1 1 1 0 1 1 0 0 1 1 1 0
[149] 1 1 1 1 0 0 0 0 1 1 1 1 1 1 0 1 1 0 1 1 1 1 1 0 1 1 1 1 0 1 1 1 0 1 0 0 1
[186] 1 1 1 0 0 1 1 1 0 1 1 0 1 0 0 1 1 1 1 1 1 1 1 1 0 0 1 1 1 0 0 1 0 1 0 1 1
[223] 0 1 0 1 1 1 1 1 1
We can evaluate the model using functions from the carret package.
library(caret)
test_val=data_test[9]
test_val=as.vector(test_val)
print(caret::confusionMatrix(as.factor(t(data_test[,9])),as.factor(test_predict)))
Confusion Matrix and Statistics
Reference
Prediction 0 1
0 63 87
1 3 78
Accuracy : 0.6104
95% CI : (0.5442, 0.6737)
No Information Rate : 0.7143
P-Value [Acc > NIR] : 0.9997
Kappa : 0.3092
Mcnemar's Test P-Value : <2e-16
Sensitivity : 0.9545
Specificity : 0.4727
Pos Pred Value : 0.4200
Neg Pred Value : 0.9630
Prevalence : 0.2857
Detection Rate : 0.2727
Detection Prevalence : 0.6494
Balanced Accuracy : 0.7136
'Positive' Class : 0
```
You've completed the tutorial of the package functions.
Thank you for doing so !
yanisb3812/DyrRegLog documentation built on Dec. 23, 2021, 7:15 p.m.
DyrRegLog
Installing the package
To use the package, it has to be installed first.
devtools::install_github("yanisb3812/DyrRegLog")
Loading the library
When the package is installed, it can be loaded by using the library command from R.
library(DyrRegLog)
Dataset Import
A dataset is required to run the functions of this package. We'll be using the diabetes2 dataset.
data<-("diabetes2.csv")
Then we can use some commands from R to visualize our data.
str(data)
data.frame': 768 obs. of 9 variables:
$ Pregnancies : int 6 1 8 1 0 5 3 10 2 8 ...
$ Glucose : int 148 85 183 89 137 116 78 115 197 125 ...
$ BloodPressure : int 72 66 64 66 40 74 50 0 70 96 ...
$ SkinThickness : int 35 29 0 23 35 0 32 0 45 0 ...
$ Insulin : int 0 0 0 94 168 0 88 0 543 0 ...
$ BMI : num 33.6 26.6 23.3 28.1 43.1 25.6 31 35.3 30.5 0 ...
$ DiabetesPedigreeFunction: num 0.627 0.351 0.672 0.167 2.288 ...
$ Age : int 50 31 32 21 33 30 26 29 53 54 ...
$ Outcome : int 1 0 1 0 1 0 1 0 1 1 ...
Now we can detect our target variable and we see that all the variables are numeric.
It includes variables that can be the cause of the presence of diabetes, shown by the target variable "Outcome".
summary(data)
Pregnancies Glucose BloodPressure SkinThickness Insulin BMI DiabetesPedigreeFunction Age Outcome
Min. : 0.000 Min. : 0.0 Min. : 0.00 Min. : 0.00 Min. : 0.0 Min. : 0.00 Min. :0.0780 Min. :21.00 Min. :0.000
1st Qu.: 1.000 1st Qu.: 99.0 1st Qu.: 62.00 1st Qu.: 0.00 1st Qu.: 0.0 1st Qu.:27.30 1st Qu.:0.2437 1st Qu.:24.00 1st Qu.:0.000
Median : 3.000 Median :117.0 Median : 72.00 Median :23.00 Median : 30.5 Median :32.00 Median :0.3725 Median :29.00 Median :0.000
Mean : 3.845 Mean :120.9 Mean : 69.11 Mean :20.54 Mean : 79.8 Mean :31.99 Mean :0.4719 Mean :33.24 Mean :0.349
3rd Qu.: 6.000 3rd Qu.:140.2 3rd Qu.: 80.00 3rd Qu.:32.00 3rd Qu.:127.2 3rd Qu.:36.60 3rd Qu.:0.6262 3rd Qu.:41.00 3rd Qu.:1.000
Max. :17.000 Max. :199.0 Max. :122.00 Max. :99.00 Max. :846.0 Max. :67.10 Max. :2.4200 Max. :81.00 Max. :1.000
We can see that our variables are not on the same scale, we'll resolve that later on.
```
After that, we'll define a size of sample.
```
smp_size <- floor(0.70 * nrow(data))
```
Here, we selected 70% of the data size.
Then, we divide our sample into a training data and a test data randomly.
```
set.seed(29112021)
train_ind <- sample(seq_len(nrow(data)), size = smp_size)
data_train <- data[train_ind, ]
data_test <- data[-train_ind, ]
```
Now we can call the fit function of DyrRegLog package to fit our model to a Logistic Regression using the stochastic gradient descent algorithm.
test_fit = fit(formula = Outcome~. , data = data , eta = 0.3 , iter_Max = 10000 , mode = "Batch_Simple" , batch_size = 20, nb_Coeurs = 1 )
We can also visualize the plot of the deviances.

The print and summary functions are overridden.
So, if we print the variable containing the fit function, it will display the formula and the coefficients.
print(test_fit)
Formula : ~ Outcome .
Coefficients : Pregnancies Glucose BloodPressure SkinThickness Insulin BMI DiabetesPedigreeFunction Age Intercept 1 0.4080121 1.110398 -0.2523509 0.005541574 -0.1286005 0.7017453 0.3108529 0.1803396 -0.8660777
It is the same thing for the summary function, but it will display the last value of
the deviance, the number of tierations and the time of execution of the algorithm.
summary(test_fit)
Last deviance value : 723.4693
Number of iterations : 180
Computing time : 0.35
Then we can predict our target variable with the predict function.
test_predict=predict(data_test[,-9], test_fit,sortie="Class") print(test_predict) [1] 0 1 0 0 1 0 1 1 1 1 1 1 1 1 1 1 1 1 0 1 1 0 1 1 0 1 0 1 0 1 1 1 0 1 0 0 1 [38] 1 0 1 0 1 0 1 0 1 1 0 0 1 1 1 1 1 0 1 1 1 1 0 1 1 1 1 1 0 1 1 0 1 1 1 1 1 [75] 1 1 0 0 1 1 0 0 1 1 1 1 1 0 1 1 1 0 1 1 1 1 1 0 1 1 1 1 1 1 1 1 1 1 0 1 1 [112] 1 1 1 1 1 1 0 0 1 0 1 1 1 0 1 1 1 1 1 0 1 1 0 0 1 1 1 1 0 1 1 0 0 1 1 1 0 [149] 1 1 1 1 0 0 0 0 1 1 1 1 1 1 0 1 1 0 1 1 1 1 1 0 1 1 1 1 0 1 1 1 0 1 0 0 1 [186] 1 1 1 0 0 1 1 1 0 1 1 0 1 0 0 1 1 1 1 1 1 1 1 1 0 0 1 1 1 0 0 1 0 1 0 1 1 [223] 0 1 0 1 1 1 1 1 1
We can evaluate the model using functions from the carret package.
library(caret) test_val=data_test[9] test_val=as.vector(test_val)
print(caret::confusionMatrix(as.factor(t(data_test[,9])),as.factor(test_predict))) Confusion Matrix and Statistics
Reference
Prediction 0 1 0 63 87 1 3 78
Accuracy : 0.6104
95% CI : (0.5442, 0.6737)
No Information Rate : 0.7143
P-Value [Acc > NIR] : 0.9997
Kappa : 0.3092
Mcnemar's Test P-Value : <2e-16
Sensitivity : 0.9545
Specificity : 0.4727
Pos Pred Value : 0.4200
Neg Pred Value : 0.9630
Prevalence : 0.2857
Detection Rate : 0.2727
Detection Prevalence : 0.6494 Balanced Accuracy : 0.7136
'Positive' Class : 0
```
You've completed the tutorial of the package functions. Thank you for doing so !
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
