README.md
In rAltmetric: Retrieves altmerics data for any published paper from altmetric.com
rAltmetric
This package provides a way to programmatically retrieve altmetric data from altmetric.com for any publication with the appropriate identifer. The package is really simple to use and only has two major functions: One (altmetrics()) to download metrics and another (altmetric_data()) to extract the data into a data.frame. It also includes generic S3 methods to plot/print metrics for any altmetric object.
Questions, features requests and issues should go here. General comments to karthik.ram@gmail.com.
Installing the package
A stable version is available from CRAN. To install
install.packages('rAltmetric')
Development version
# If you don't already have the devtools library, first run
install.packages('devtools')
# then install the package
library(devtools)
install_github('rAltmetric', 'ropensci')
Quick Tutorial
Obtaining metrics
There was a recent paper by Acuna et al that received a lot of attention on Twitter. What was the impact of that paper?
library(rAltmetric)
acuna <- altmetrics('10.1038/489201a')
> acuna
Altmetrics on: "Future impact: Predicting scientific success" with doi 10.1038/489201a (altmetric_id: 942310) published in Nature.
provider count
1 Feeds 9
2 Google+ 1
3 Cited 174
4 Tweets 157
5 Accounts 167
Data
To obtain the metrics in tabular form for further processing, run any object of class altmetric through altmetric_data() to get data that can easily be written to disk as a spreadsheet.
> altmetric_data(acuna)
title
1 Future impact: Predicting scientific success
doi nlmid altmetric_jid issns
1 10.1038/489201a 0410462 4f6fa50a3cf058f610003160 0028-0836
journal altmetric_id schema is_oa cited_by_feeds_count
1 Nature 942310 1.5.4 FALSE 173
cited_by_gplus_count cited_by_posts_count
1 173 173
cited_by_tweeters_count cited_by_accounts_count score
1 156 166 184.598
mendeley connotea citeulike pub sci com doc
1 0 0 11 62 84 6 8
url
1 http://www.nature.com/nature/journal/v489/n7415/full/489201a.html
added_on published_on subjects scopus_subjects
1 1347471425 1347404400 science General
last_updated readers_count X1 count_all count_journal
1 1348828350 11 1 754555 13972
count_similar_age_1m count_similar_age_3m
1 22408 56213
count_similar_age_journal_1m count_similar_age_journal_3m
1 508 1035
rank_all rank_journal rank_similar_age_1m
1 754043 13759 22339
rank_similar_age_3m rank_similar_age_journal_1m
1 56074 459
rank_similar_age_journal_3m pct_all pct_journal
1 947 99.93 98.48
pct_similar_age_1m pct_similar_age_3m
1 99.69 99.75
pct_similar_age_journal_1m pct_similar_age_journal_3m
1 90.35 91.50
details_url
1 http://www.altmetric.com/details.php?citation_id=942310
You can save these data into a clean spreadsheet format:
acuna_data <- altmetric_data(acuna)
write.csv(acuna_data, file = 'acuna_altmetrics.csv')
Visualization
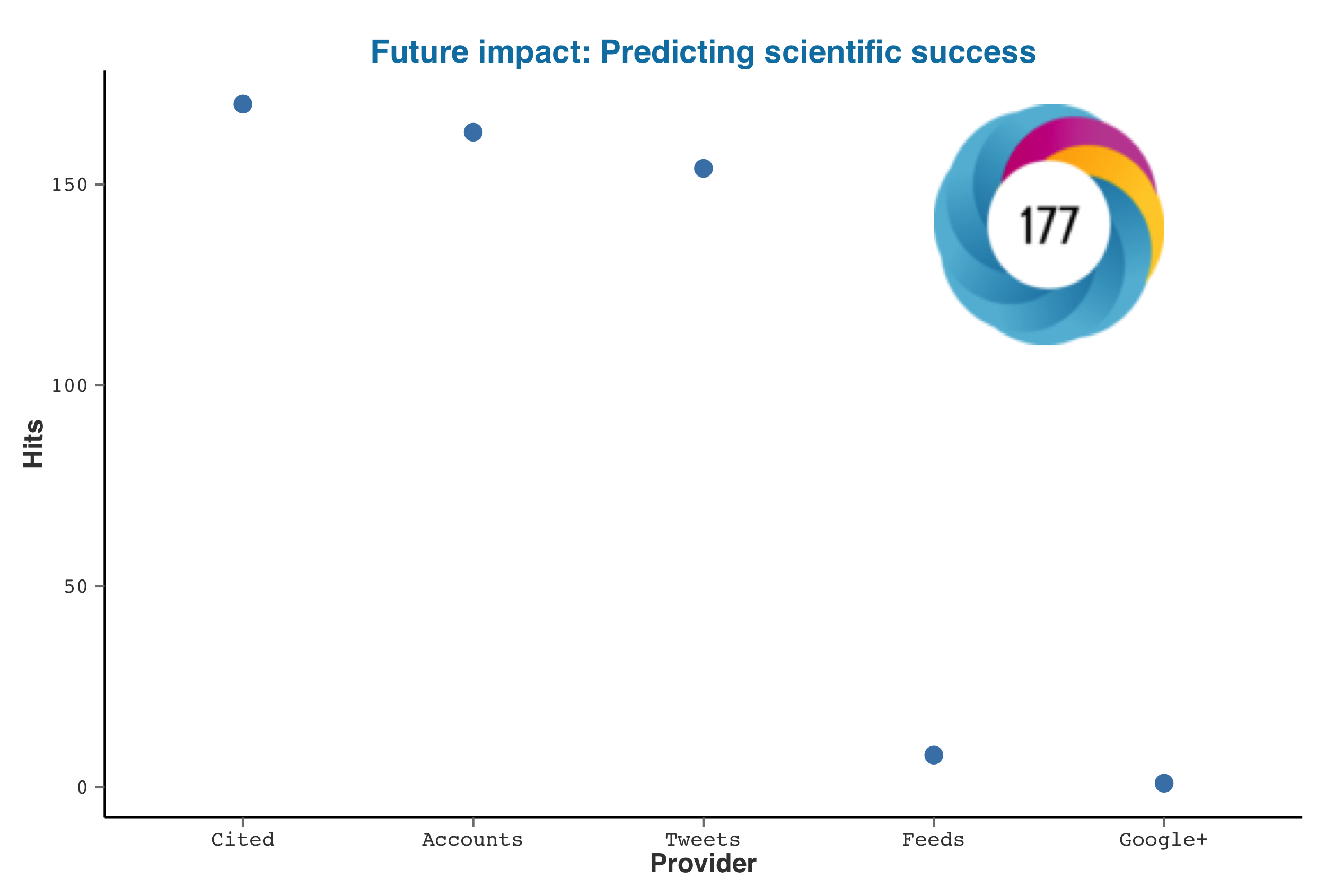
For any altmetric object you can quickly plot the stats with a generic plot function. The plot overlays the altmetric badge and the score on the top right corner. If you prefer a customized plot, create your own with the raw data generated from almetric_data()
> plot(acuna)
Gathering metrics for many DOIs
For a real world use-case, one might want to get metrics on multiple publications. If so, just read them from a spreadsheet and llply through them like the example below.
# Be sure to update the path if the example csv is not in your working dir
doi_data <- read.csv('dois.csv', header = TRUE)
> doi_data
doi
1 10.1038/nature09210
2 10.1126/science.1187820
3 10.1016/j.tree.2011.01.009
4 10.1086/664183
library(plyr)
# First, let's retrieve the metrics.
raw_metrics <- llply(doi_data$doi, function(x) altmetrics(doi = x), .progress = 'text')
# Now let's pull the data together.
metric_data <- ldply(raw_metrics, altmetric_data)
# Finally we save this to a spreadsheet for further analysis/vizualization.
write.csv(metric_data, file = "metric_data.csv")
Further reading
Try the rAltmetric package in your browser
Any scripts or data that you put into this service are public.
rAltmetric documentation built on May 2, 2019, 4:22 p.m.

rAltmetric
This package provides a way to programmatically retrieve altmetric data from altmetric.com for any publication with the appropriate identifer. The package is really simple to use and only has two major functions: One (altmetrics()) to download metrics and another (altmetric_data()) to extract the data into a data.frame. It also includes generic S3 methods to plot/print metrics for any altmetric object.
Questions, features requests and issues should go here. General comments to karthik.ram@gmail.com.
Installing the package
A stable version is available from CRAN. To install
install.packages('rAltmetric')
Development version
# If you don't already have the devtools library, first run
install.packages('devtools')
# then install the package
library(devtools)
install_github('rAltmetric', 'ropensci')
Quick Tutorial
Obtaining metrics
There was a recent paper by Acuna et al that received a lot of attention on Twitter. What was the impact of that paper?
library(rAltmetric)
acuna <- altmetrics('10.1038/489201a')
> acuna
Altmetrics on: "Future impact: Predicting scientific success" with doi 10.1038/489201a (altmetric_id: 942310) published in Nature.
provider count
1 Feeds 9
2 Google+ 1
3 Cited 174
4 Tweets 157
5 Accounts 167
Data
To obtain the metrics in tabular form for further processing, run any object of class altmetric through altmetric_data() to get data that can easily be written to disk as a spreadsheet.
> altmetric_data(acuna)
title
1 Future impact: Predicting scientific success
doi nlmid altmetric_jid issns
1 10.1038/489201a 0410462 4f6fa50a3cf058f610003160 0028-0836
journal altmetric_id schema is_oa cited_by_feeds_count
1 Nature 942310 1.5.4 FALSE 173
cited_by_gplus_count cited_by_posts_count
1 173 173
cited_by_tweeters_count cited_by_accounts_count score
1 156 166 184.598
mendeley connotea citeulike pub sci com doc
1 0 0 11 62 84 6 8
url
1 http://www.nature.com/nature/journal/v489/n7415/full/489201a.html
added_on published_on subjects scopus_subjects
1 1347471425 1347404400 science General
last_updated readers_count X1 count_all count_journal
1 1348828350 11 1 754555 13972
count_similar_age_1m count_similar_age_3m
1 22408 56213
count_similar_age_journal_1m count_similar_age_journal_3m
1 508 1035
rank_all rank_journal rank_similar_age_1m
1 754043 13759 22339
rank_similar_age_3m rank_similar_age_journal_1m
1 56074 459
rank_similar_age_journal_3m pct_all pct_journal
1 947 99.93 98.48
pct_similar_age_1m pct_similar_age_3m
1 99.69 99.75
pct_similar_age_journal_1m pct_similar_age_journal_3m
1 90.35 91.50
details_url
1 http://www.altmetric.com/details.php?citation_id=942310
You can save these data into a clean spreadsheet format:
acuna_data <- altmetric_data(acuna)
write.csv(acuna_data, file = 'acuna_altmetrics.csv')
Visualization
For any altmetric object you can quickly plot the stats with a generic plot function. The plot overlays the altmetric badge and the score on the top right corner. If you prefer a customized plot, create your own with the raw data generated from almetric_data()
> plot(acuna)

Gathering metrics for many DOIs
For a real world use-case, one might want to get metrics on multiple publications. If so, just read them from a spreadsheet and llply through them like the example below.
# Be sure to update the path if the example csv is not in your working dir
doi_data <- read.csv('dois.csv', header = TRUE)
> doi_data
doi
1 10.1038/nature09210
2 10.1126/science.1187820
3 10.1016/j.tree.2011.01.009
4 10.1086/664183
library(plyr)
# First, let's retrieve the metrics.
raw_metrics <- llply(doi_data$doi, function(x) altmetrics(doi = x), .progress = 'text')
# Now let's pull the data together.
metric_data <- ldply(raw_metrics, altmetric_data)
# Finally we save this to a spreadsheet for further analysis/vizualization.
write.csv(metric_data, file = "metric_data.csv")
Further reading
Try the rAltmetric package in your browser
Any scripts or data that you put into this service are public.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
