README.md
In ctrdata: Retrieve and Analyze Clinical Trials Data from Public Registers
Main features • References •
Installation •
Overview •
Databases • Data
model • Example workflow •
Analysis across registers •
Tests • Acknowledgements •
Future
ctrdata for aggregating and analysing clinical trials
The package ctrdata provides functions for retrieving (downloading),
aggregating and analysing clinical trials using information (structured
protocol and result data, as well as documents) from public registers.
It can be used with the
- EU Clinical Trials Register (“EUCTR”,
https://www.clinicaltrialsregister.eu/)
- EU Clinical Trials Information System (“CTIS”,
https://euclinicaltrials.eu/, see example)
- ClinicalTrials.gov (“CTGOV2”, see example)
- ISRCTN Registry (https://www.isrctn.com/)
The motivation is to investigate the design and conduct of trials of
interest, to describe their trends and availability for patients and to
facilitate using their detailed results for research and meta-analyses.
ctrdata is a package for the R system,
but other systems and tools can use the databases created with this
package. This README was reviewed on 2025-06-07 for version 1.22.2.9000.
Main features
-
Protocol- and results-related trial information is easily downloaded:
Users define a query in a register’s web interface, then copy the URL
and enter it into ctrdata which retrieves in one go all trials
found. A
script
can automate copying the query URL from all registers. Personal
annotations can be made when downloading trials. Also, trial
documents and historic
versions
as available in registers on trials can be downloaded.
-
Downloaded trial information is transformed and stored in a collection
of a document-centric database, for fast and offline access.
Information from different registers can be accumulated in a single
collection. Uses DuckDB, PostgreSQL, RSQLite or MongoDB, via R
package nodbi: see section
Databases below.
Interactively browse through trial structure and data. Easily re-run
any previous query in a collection to retrieve and update trial
records.
-
For analyses, convenience functions in ctrdata implement canonical
trial
concepts
to simplify analyses across registers 🔔, allow find synonyms of an
active substance, identify unique (de-duplicated) trial records across
all registers, to merge and recode fields as well as to easily access
deeply-nested fields. Analysis can be done with R (see
vignette)
or other systems, using the JSON-structured information in the
database.
Remember to respect the registers’ terms and conditions (see
ctrOpenSearchPagesInBrowser(copyright = TRUE)). Please cite this
package in any publication as follows: “Ralf Herold (2025). ctrdata:
Retrieve and Analyze Clinical Trials in Public Registers. R package
version 1.22.2, https://cran.r-project.org/package=ctrdata”.
References
Package ctrdata has been used for unpublished works and these
publications:
- Jong et al. (2025) Experiences with Low-Intervention Clinical
Trials—the New Category under the European Union Clinical Trials
Regulation. Clinical Trials
https://doi.org/10.1177/17407745241309293
- Lopez-Rey et al. (2025) Use of Bayesian Approaches in Oncology
Clinical Trials: A Cross-Sectional Analysis’. Frontiers in
Pharmacology https://doi.org/10.3389/fphar.2025.1548997
- Russek et al. (2025) Supplementing Single-Arm Trials with External
Control Arms—Evaluation of German Real-World Data. Clinical
Pharmacology & Therapeutics https://doi.org/10.1002/cpt.3684
- Alzheimer’s disease Horizon Scanning Report (2024) PDF file, p
10
- Kundu et al. (2024) Analysis of Factors Influencing Enrollment Success
in Hematology Malignancy Cancer Clinical Trials (2008-2023). Blood
Meeting Abstracts https://doi.org/10.1182/blood-2024-207446
- Lasch et al. (2022) The Impact of COVID‐19 on the Initiation of
Clinical Trials in Europe and the United States. Clinical Pharmacology
& Therapeutics https://doi.org/10.1002/cpt.2534
- Sood et al. (2022) Managing the Evidence Infodemic: Automation
Approaches Used for Developing NICE COVID-19 Living Guidelines.
medRxiv https://doi.org/10.1101/2022.06.13.22276242
- Blogging on Innovation coming to paediatric
research
- Cancer Research UK (2017) The impact of collaboration: The value of
UK medical research to EU science and
health
- EMA (2017) Results of juvenile animal studies (JAS) and impact on
anti-cancer medicine development and use in children PDF file, p
34
Installation
1. Install package ctrdata in R
Package ctrdata is on
CRAN and on
GitHub. Within
R, use the following commands to install
package ctrdata:
# Install CRAN version:
install.packages("ctrdata")
# Alternatively, install development version:
install.packages("devtools")
devtools::install_github("rfhb/ctrdata", build_vignettes = TRUE)
These commands also install the package’s dependencies (jsonlite,
httr, curl, clipr, xml2, nodbi, stringi, tibble,
lubridate, jqr, dplyr, zip, readr, digest, countrycode,
htmlwidgets, stringdist and V8).
2. Script to automatically copy user’s query from web browser
Optional; works with all registers supported by ctrdata and is
recommended for CTIS so that its URL in the web browser reflects the
user’s parameters for querying this register.
In the web browser, install the Tampermonkey browser
extension, click on “New user script”
and then on “Tools”, enter into “Import from URL” this URL:
https://raw.githubusercontent.com/rfhb/ctrdata/master/tools/ctrdataURLcopier.js
and then click on “Install”.
The browser extension can be disabled and enabled by the user. When
enabled, the URLs to all user’s queries in the registers are
automatically copied to the clipboard and can be pasted into the
queryterm = ... parameter of function
ctrLoadQueryIntoDb().
Additionally, this script retrieves results for CTIS when opening
search URLs such as
https://euclinicaltrials.eu/ctis-public/search#searchCriteria={"status":[3,4]}.
After changing the URL in the browser, a “Reload page” is needed to
conduct the search and show results.
Overview of functions in ctrdata
The functions are listed in the approximate order of use in a user’s
workflow (in bold, main functions). See also the package documentation
overview.
| Function name | Function purpose |
|----|----|
| ctrOpenSearchPagesInBrowser() | Open search pages of registers or execute search in web browser |
| ctrFindActiveSubstanceSynonyms() | Find synonyms and alternative names for an active substance |
| ctrGenerateQueries() | 🔔 From simple user parameters, generates queries for each register to find trials of interest |
| ctrGetQueryUrl() | Import from clipboard the URL of a search in one of the registers |
| ctrLoadQueryIntoDb() | Retrieve (download) or update, and annotate, information on trials from a register and store in a collection in a database |
| ctrShowOneTrial() | 🔔 Show full structure and all data of a trial, interactively select fields of interest for dbGetFieldsIntoDf() |
| dbQueryHistory() | Show the history of queries that were downloaded into the collection |
| dbFindIdsUniqueTrials() | Get the identifiers of de-duplicated trials in the collection |
| dbFindFields() | Find names of variables (fields) in the collection |
| dbGetFieldsIntoDf() | Create a data frame (or tibble) from trial records in the database with the specified fields |
| dfTrials2Long() | Transform the data.frame from dbGetFieldsIntoDf() into a long name-value data.frame, including deeply nested fields |
| dfName2Value() | From a long name-value data.frame, extract values for variables (fields) of interest (e.g., endpoints) |
| dfMergeVariablesRelevel() | Merge variables into a new variable, optionally map values to a new set of levels |
Databases for use with ctrdata
Package ctrdata retrieves trial information and stores it in a
database collection, which has to be given as a connection object to
parameter con for several ctrdata functions. This connection object
is created almost identically for the four database backends supported
by ctrdata, as shown in the table. For a speed comparison, see the
nodbi documentation.
Besides ctrdata functions below, such a connection object can be used
with functions of other packages, such as nodbi (see last row in
table) or, in case of MongoDB as database backend, mongolite (see
vignettes).
| Purpose | Function call |
|----|----|
| Create SQLite database connection | dbc <- nodbi::src_sqlite(dbname = "name_of_my_database", collection = "name_of_my_collection") |
| Create DuckDB database connection | dbc <- nodbi::src_duckdb(dbdir = "name_of_my_database", collection = "name_of_my_collection") |
| Create MongoDB database connection | dbc <- nodbi::src_mongo(db = "name_of_my_database", collection = "name_of_my_collection") |
| Create PostgreSQL database connection | dbc <- nodbi::src_postgres(dbname = "name_of_my_database"); dbc[["collection"]] <- "name_of_my_collection" |
| Use connection with ctrdata functions | ctrdata::{ctrLoadQueryIntoDb, dbQueryHistory, dbFindIdsUniqueTrials, dbFindFields, dbGetFieldsIntoDf}(con = dbc, ...) |
| Use connection with nodbi functions | e.g., nodbi::docdb_query(src = dbc, key = dbc$collection, ...) |
Data model of ctrdata
Package ctrdata uses the data models that are implicit in data as
retrieved from the different registers. No mapping is provided for any
register’s data model to a putative target data model. The reasons
include that registers’ data models are continually evolving over time,
that only few data fields have similar values and meaning between
registers, and that the retrieved public data may not correspond to the
registers’ internal data model. The structure of data for a specific
trial can interactively be inspected and searched using function, see
the section below.
Thus, the handling of data from different models of registers is to be
done at the time of analysis. This approach allows a high level of
flexibility, transparency and reproducibility. To support analyses,
ctrdata (from version 1.21.0) provides functions that calculate
concepts of clinical trials across registers, which are commonly used in
analyses, such as start dates, age groups and statistical tests of
results. See
help(ctrdata-trial-concepts)
and the section Analysis across
trials in the example workflow
below. For further analyses, see examples of function
dfMergeVariablesRelevel()
on how to align related fields from different registers for a joint
analysis.
In any of the
databases,
one clinical trial is one document, corresponding to one row in a
SQLite, PostgreSQL or DuckDB table, and to one document in a
MongoDB collection. These NoSQL backends allow documents to have
different structures, which is used here to accommodate the different
models of data retrieved from the registers. Package ctrdata stores in
every such document:
- field
_id with the trial identification as provided by the register
from which it was retrieved
- field
ctrname with the name of the register (EUCTR, CTGOV,
CTGOV2, ISRCTN, CTIS) from which that trial was retrieved
- field
record_last_import with the date and time when that document
was last updated using ctrLoadQueryIntoDb()
- only for
CTGOV2 and CTIS: object history with a historic version
of the trial and with history_version, which contains the fields
version_number (starting from 1) and version_date
- all original fields as provided by the register for that trial (see
example in
vignette)
For visualising the data structure for a trial, see this vignette
section.
Vignettes
- Install R package
ctrdata
- Retrieve clinical trial
information
- Summarise and analyse clinical trial
information
Example workflow
The aim is to download protocol-related trial information and tabulate
the trials’ status of conduct.
- Attach package
ctrdata:
library(ctrdata)
- See help to get started with
ctrdata:
help("ctrdata")
- Information on trial registers, their contents and how they can be
used with
ctrdata (last updated 2025-05-31):
help("ctrdata-registers")
- Trial concepts across registers provided by
ctrdata (new 2025-03-09
🔔):
help("ctrdata-trial-concepts")
- Open registers’ advanced search pages in browser:
ctrOpenSearchPagesInBrowser()
# Please review and respect register copyrights:
ctrOpenSearchPagesInBrowser(copyright = TRUE)
-
Adjust search parameters and execute search in browser
-
When trials of interest are listed in browser, copy the address from
the browser’s address bar to the clipboard (you can automate this,
see
here)
-
Search used in this example:
https://www.clinicaltrialsregister.eu/ctr-search/search?query=neuroblastoma&phase=phase-two&age=children
-
Get address from clipboard:
q <- ctrGetQueryUrl()
# * Using clipboard content as register query URL: https://www.clinicaltrialsregister.eu/
# ctr-search/search?query=neuroblastoma&phase=phase-two&age=children
# * Found search query from EUCTR: query=neuroblastoma&phase=phase-two&age=children
q
# query-term query-register
# 1 query=neuroblastoma&phase=phase-two&age=children EUCTR
🔔 Queries in the trial registers can automatically copied to the
clipboard (including for “CTIS”, where the URL otherwise does not show
the user’s query) using the solution
here.
- Retrieve protocol-related information, transform and save to database:
For loading the trial information, first a database collection is
specified, using nodbi (see above for how to specify PostgreSQL,
RSQlite, DuckDB or MongoDB as backend, see section
Databases):
# Connect to (or create) an SQLite database
# stored in a file on the local system:
db <- nodbi::src_sqlite(
dbname = "database_name.sql",
collection = "collection_name"
)
Then, the trial information is retrieved and loaded into the collection:
# Retrieve trials from public register:
ctrLoadQueryIntoDb(
queryterm = q,
euctrresults = TRUE,
con = db
)
# * Checking trials in EUCTR...
# Retrieved overview, multiple records of 73 trial(s) from 4 page(s) to be downloaded (estimate: 9 MB)
# (1/3) Downloading trials...
# Note: register server cannot compress data, transfer takes longer (estimate: 90 s)
# Download status: 4 done; 0 in progress. Total size: 6.39 Mb (100%)... done!
# (2/3) Converting to NDJSON (estimate: 1 s)...
# (3/3) Importing records into database...
# = Imported or updated 270 records on 73 trial(s)
# * Checking results if available from EUCTR for 73 trials:
# (1/4) Downloading results...
# Download status: 73 done; 0 in progress. Total size: 22.57 Mb (100%)... done!
# Download status: 41 done; 0 in progress. Total size: 165.04 Kb (100%)... done!
# Download status: 41 done; 0 in progress. Total size: 165.04 Kb (100%)... done!
# - extracting results (. = data, F = file[s] and data, x = none):
# F F . . . . . . . F . . . F . . . F . . . . . F . . . . . . . F
# (2/4) Converting to NDJSON (estimate: 3 s)...
# (3/4) Importing results into database (may take some time)...
# (4/4) Results history: not retrieved (euctrresultshistory = FALSE)
# = Imported or updated results for 32 trials
# No history found in expected format.
# Updated history ("meta-info" in "some_collection_name")
# $n
# [1] 270
Under the hood, EUCTR plain text and XML files from EUCTR, CTGOV, ISRCTN
are converted using Javascript via V8 in R into NDJSON, which is
imported into the database collection.
- Easily generate queries for each register and add records from several
registers into the same collection
The same parameters can be used to ask ctrdata to generate search
queries that apply to each register, for opening the web interfaces and
for loading the trial data into the collection:
# Generate queries for each register
queries <- ctrGenerateQueries(
condition = "neuroblastoma",
recruitment = "completed",
phase = "phase 2",
population = "P"
)
queries
# EUCTR
# "https://www.clinicaltrialsregister.eu/ctr-search/search?query=neuroblastoma&phase=phase-two&age=children&age=adolescent&age=infant-and-toddler&age=newborn&age=preterm-new-born-infants&age=under-18&status=completed"
# ISRCTN
# "https://www.isrctn.com/search?&q=&filters=condition:neuroblastoma,phase:Phase II,ageRange:Child,trialStatus:completed,primaryStudyDesign:Interventional"
# CTGOV2
# "https://clinicaltrials.gov/search?cond=neuroblastoma&aggFilters=phase:2,ages:child,status:com,studyType:int"
# CTGOV2expert
# "https://clinicaltrials.gov/expert-search?term=AREA[ConditionSearch]\"neuroblastoma\" AND (AREA[Phase]\"PHASE2\") AND (AREA[StdAge]\"CHILD\") AND (AREA[OverallStatus]\"COMPLETED\") AND (AREA[StudyType]INTERVENTIONAL)"
# CTIS
# "https://euclinicaltrials.eu/ctis-public/search#searchCriteria={\"medicalCondition\":\"neuroblastoma\",\"trialPhaseCode\":[4],\"ageGroupCode\":[2],\"status\":[5,8]}"
# Open queries in registers' web interfaces
sapply(queries, ctrOpenSearchPagesInBrowser)
# Load all queries into database collection
result <- lapply(queries, ctrLoadQueryIntoDb, con = db)
sapply(result, "[[", "n")
# EUCTR ISRCTN CTGOV2 CTGOV2expert CTIS
# 180 0 110 110 1
- Analyse
Tabulate the status of trials that are part of an agreed paediatric
development program (paediatric investigation plan, PIP). ctrdata
functions return a data.frame (or a tibble, if package tibble is
loaded).
# Get all records that have values in the fields of interest:
result <- dbGetFieldsIntoDf(
# Field of interest
fields = c("a7_trial_is_part_of_a_paediatric_investigation_plan"),
# Trial concepts calculated across registers
calculate = c("f.statusRecruitment", "f.isUniqueTrial"),
con = db
)
# Querying database (16 fields)...
# Searching for duplicate trials...
# - Getting all trial identifiers (may take some time), 381 found in collection
# - Finding duplicates among registers' and sponsor ids...
# - 174 EUCTR _id were not preferred EU Member State record for 66 trials
# - Keeping 110 / 60 / 0 / 0 / 0 records from CTGOV2 / EUCTR / CTGOV / ISRCTN / CTIS
# = Returning keys (_id) of 170 records in collection "collection_name"
# Tabulate the clinical trial information of interest
with(
result[result$.isUniqueTrial, ],
table(
.statusRecruitment,
a7_trial_is_part_of_a_paediatric_investigation_plan
)
)
# a7_trial_is_part_of_a_paediatric_investigation_plan
# .statusRecruitment FALSE TRUE
# ongoing 2 3
# completed 12 5
# ended early 7 3
# other 9 3
- Queries to CTGOV and CTGOV2
The new website and API introduced in July 2023
(https://www.clinicaltrials.gov/) is supported by ctrdata since
mid-2023 and identified in ctrdata as CTGOV2.
On 2024-06-25, CTGOV has retired the classic website and API used by
ctrdata since 2015. To support users, ctrdata automatically
translates and redirects queries to the current website. This helps with
automatically updating previously loaded queries
(ctrLoadQueryIntoDb(querytoupdate = <n>)), manually migrating queries
and reproducible work on clinical trials information. Going forward,
users are recommended to change to use CTGOV2 queries.
As regards study data, important differences exist between field names
and contents of information retrieved using CTGOV or CTGOV2; see the
schema for study protocols in
CTGOV,
the schema for study
results
and the Study Data Structure for
CTGOV2.
For more details, call help("ctrdata-registers"). This is one of the
reasons why ctrdata handles the situation as if these were two
different registers and will continue to identify the current API as
register = "CTGOV2", to support the analysis stage.
Note that loading trials with ctrdata overwrites the previous record
with CTGOV2 data, whether the previous record was retrieved using
CTGOV or CTGOV2 queries.
Example using a CTGOV query:
# CTGOV search query URL
q <- "https://classic.clinicaltrials.gov/ct2/results?cond=neuroblastoma&rslt=With&recrs=e&age=0&intr=Drug"
# Open old URL (CTGOV) in current website (CTGOV2):
ctrOpenSearchPagesInBrowser(q)
# * Appears specific for CTGOV Classic website
# Since 2024-06-25, the classic CTGOV servers are no longer available.
# Package ctrdata has translated the classic CTGOV query URL from this call
# of function ctrLoadQueryIntoDb(queryterm = ...) into a query URL that works
# with the current CTGOV2. This is printed below and is also part of the return
# value of this function, ctrLoadQueryIntoDb(...)$url. This URL can be used with
# ctrdata functions. Note that the fields and data schema of trials differ
# between CTGOV and CTGOV2.
#
# Replace this URL:
#
# https://classic.clinicaltrials.gov/ct2/results?cond=neuroblastoma&rslt=With&recrs=e&age=0&intr=Drug
#
# with this URL:
#
# https://clinicaltrials.gov/search?cond=neuroblastoma&intr=Drug&aggFilters=ages:child,results:with,status:com
#
# * Found search query from CTGOV2: cond=neuroblastoma&intr=Drug&aggFilters=ages:child,results:with,status:com
# Count trials:
ctrLoadQueryIntoDb(
queryterm = q,
con = db,
only.count = TRUE
)
# $n
# [1] 71
- Queries to CTIS
Queries in the CTIS search interface can be automatically copied to the
clipboard so that a user can paste them into queryterm, see
here.
Subsequent to the relaunch of CTIS on 2024-07-18, there are more than
8,700 trials publicly accessible in CTIS. See
below for how to download documents from CTIS.
# See how many trials are in CTIS publicly accessible:
ctrLoadQueryIntoDb(
queryterm = "",
register = "CTIS",
only.count = TRUE
)
# $n
# [1] 9181
- Inspect and search structure of trial information
For a given trial, function
ctrShowOneTrial()
enables the user to visualise the hiearchy of fields and contents in the
user’s local web browser, to search for field names and field values,
and to select and copy selected fields’ names for use with function
dbGetFieldsIntoDf().
# This opens a local browser for user interaction.
# If the trial identifier (_id) is not found in the
# specified collection, it will be retrieved from the register.
ctrShowOneTrial(
identifier = "2024-518931-12-00",
con = db
)
- Analysis across registers
Show cumulative start of trials over time. This uses the calculation of
trial concepts as available since ctrdata version 1.21.0 🔔.
# use helper package
library(dplyr)
library(ggplot2)
df <- dbGetFieldsIntoDf(
fields = "",
calculate = c("f.statusRecruitment", "f.isUniqueTrial", "f.startDate"),
con = db
)
df %>%
filter(.isUniqueTrial) %>%
ggplot() +
stat_ecdf(aes(
x = .startDate,
colour = .statusRecruitment
)) +
labs(
title = "Evolution over time of selected trials",
subtitle = "Data from EUCTR, CTIS, ISRCTN, CTGOV2",
x = "Date of start (proposed or realised)",
y = "Cumulative proportion of trials",
colour = "Current status",
caption = Sys.Date()
)
ggsave(
filename = "man/figures/README-ctrdata_across_registers.png",
width = 5, height = 3, units = "in"
)
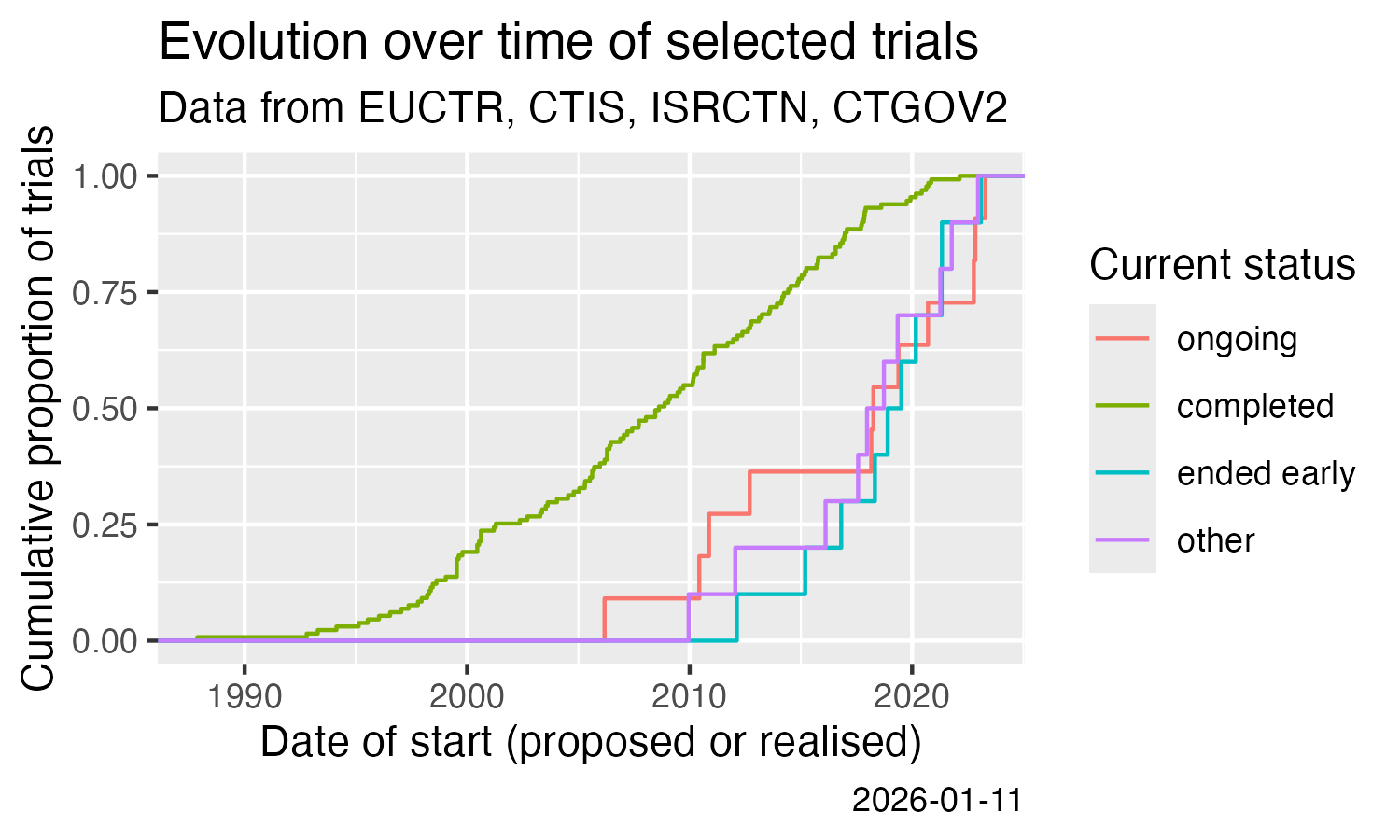 Analysis across registers
Analysis across registers
- Result-related trial information
Analyse some simple result details, here from CTGOV2 (see this
vignette
for more examples):
# use helper package
library(ggplot2)
result <- dbGetFieldsIntoDf(
calculate = c(
"f.numSites",
"f.sampleSize",
"f.controlType",
"f.numTestArmsSubstances"
),
con = db
)
ggplot(data = result) +
labs(
title = "Selected trials",
subtitle = "Patients with a neuroblastoma"
) +
geom_point(
mapping = aes(
x = .numSites,
y = .sampleSize,
size = .numTestArmsSubstances,
colour = .controlType
)
) +
scale_x_log10() +
scale_y_log10() +
labs(
x = "Number of sites",
y = "Total number of participants",
colour = "Control",
size = "# Treatments",
caption = Sys.Date()
)
ggsave(
filename = "man/figures/README-ctrdata_results_neuroblastoma.png",
width = 5, height = 3, units = "in"
)
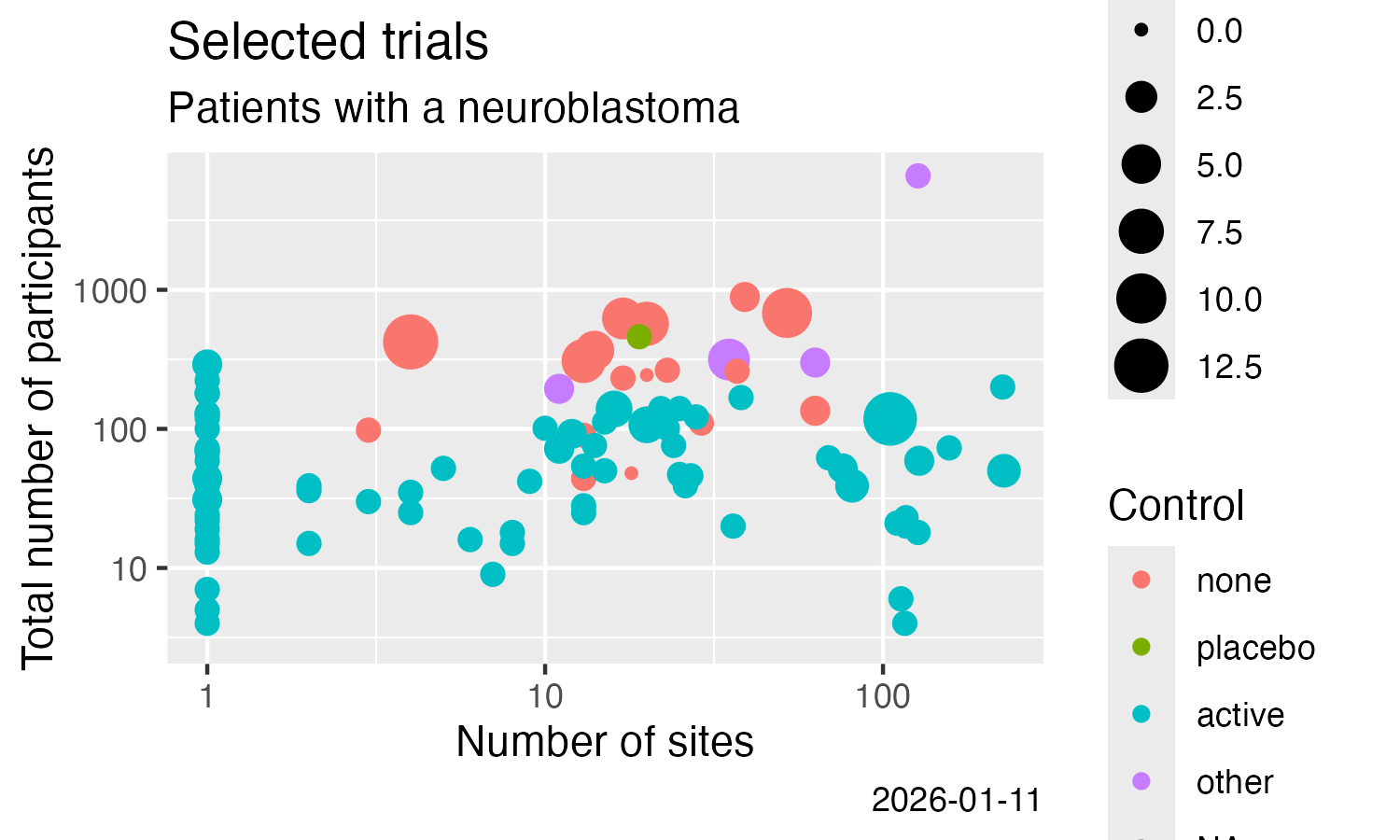 Neuroblastoma trials
Neuroblastoma trials
- Download documents: retrieve protocols, statistical analysis plans and
other documents into the local folder
./files-.../
### EUCTR document files can be downloaded when results are requested
# All files are downloaded and saved (documents.regexp is not used with EUCTR)
ctrLoadQueryIntoDb(
queryterm = "query=cancer&age=under-18&phase=phase-one",
register = "EUCTR",
euctrresults = TRUE,
documents.path = "./files-euctr/",
con = db
)
# * Found search query from EUCTR: query=cancer&age=under-18&phase=phase-one
# * Checking trials in EUCTR...
# [...]
# = documents saved in './files-euctr'
# Updated history ("meta-info" in "some_collection_name")
### CTGOV files are downloaded, here corresponding to the default of
# documents.regexp = "prot|sample|statist|sap_|p1ar|p2ars|ctalett|lay|^[0-9]+ "
ctrLoadQueryIntoDb(
queryterm = "cond=Neuroblastoma&type=Intr&recrs=e&phase=1&u_prot=Y&u_sap=Y&u_icf=Y",
register = "CTGOV",
documents.path = "./files-ctgov/",
con = db
)
# * Checking for documents...
# - Getting links to documents
# - Downloading documents into 'documents.path' = ./files-ctgov/
# - Created directory ./files-ctgov
# - Applying 'documents.regexp' to 40 missing documents
# - Creating subfolder for each trial
# - Downloading 40 missing documents
# Download status: 40 done; 0 in progress. Total size: 110.75 Mb (100%)... done!
# = Newly saved 40 document(s) for 32 trial(s); 0 of such document(s) for 0 trial(s)
# already existed in ./files-ctgov
### CTGOV2 files are downloaded, using the default of documents.regexp
ctrLoadQueryIntoDb(
queryterm = "https://clinicaltrials.gov/search?cond=neuroblastoma&aggFilters=phase:1,results:with",
documents.path = "./files-ctgov2/",
con = db
)
# * Checking for documents...
# - Getting links to documents
# - Downloading documents into 'documents.path' = ./files-ctgov2/
# - Created directory ./files-ctgov2
# - Creating subfolder for each trial
# - Applying 'documents.regexp' to 42 missing documents
# - Downloading 42 missing documents
# Download status: 42 done; 0 in progress. Total size: 92.57 Mb (100%)... done!
# = Newly saved 42 document(s) for 26 trial(s); 0 of such document(s) for 0
# trial(s) already existed in ./files-ctgov2
### ISRCTN files are downloaded, using the default of documents.regexp
ctrLoadQueryIntoDb(
queryterm = "https://www.isrctn.com/search?q=alzheimer",
documents.path = "./files-isrctn/",
con = db
)
# * Found search query from ISRCTN: q=alzheimer
# [...]
# * Checking for documents...
# - Getting links to documents
# - Downloading documents into 'documents.path' = ./files-isrctn/
# - Created directory /Users/ralfherold/Daten/mak/r/emea/ctrdata/files-isrctn
# - Applying 'documents.regexp' to 52 missing documents
# - Creating subfolder for each trial
# - Downloading 32 missing documents
# Download status: 32 done; 0 in progress. Total size: 14.89 Mb (100%)... done!
# = Newly saved 26 document(s) for 15 trial(s); 0 of such document(s) for 0
# trial(s) already existed in ./files-isrctn
### CTIS files are downloaded, using the default of documents.regexp
ctrLoadQueryIntoDb(
queryterm = paste0(
"https://euclinicaltrials.eu/ctis-public/search#",
'searchCriteria={"containAny":"cancer","status":[8]}'
),
documents.path = "./files-ctis/",
documents.regexp = "icf",
con = db
)
# * Found search query from CTIS: searchCriteria={"containAny":"cancer"}
# [...]
# * Checking for documents: . . . . . . . . . . . . . . . . . . .
# - Downloading documents into 'documents.path' = ./files-ctis/
# - Applying 'documents.regexp' to 1114 missing documents
# - Creating subfolder for each trial
# - Downloading 512 missing documents (excluding 2 files with duplicate names
# for saving, e.g. /Users/ralfherold/Daten/mak/r/emea/ctrdata/files-ctis/2022-
# 500694-14-00/SbjctInfaICF - L1 SIS and ICF Prescreening ICF clean placeholder
# - 137297.PDF, /Users/ralfherold/Daten/mak/r/emea/ctrdata/files-ctis/2022-
# 500694-14-00/SbjctInfaICF - L1 SIS and ICF Pregnant Partner ICF clean -
# 137297.PDF)
# Download status: 510 done; 0 in progress. Total size: 377.27 Kb (100%)... done!
# Redirecting to CDN...
# Download status: 127 done; 0 in progress. Total size: 47.66 Mb (100%)... done!
# = Newly saved 510 document(s) for 35 trial(s); 0 of such document(s) for 0
# trial(s) already existed in ./files-ctis
Tests and coverage
See also https://app.codecov.io/gh/rfhb/ctrdata/tree/master/R
# 2025-05-17
tinytest::test_all()
# test_ctrdata_duckdb_ctgov2.R.. 78 tests OK 46.4s
# test_ctrdata_function_activesubstance.R 4 tests OK 0.8s
# test_ctrdata_function_ctrgeneratequeries.R 14 tests OK 18ms
# test_ctrdata_function_params.R 25 tests OK 0.8s
# test_ctrdata_function_trial-concepts.R 80 tests OK 3.2s
# test_ctrdata_function_various.R 76 tests OK 3.7s
# test_ctrdata_postgres_ctgov2.R 50 tests OK 32.4s
# test_ctrdata_sqlite_ctgov.R... 46 tests OK 29.0s
# test_ctrdata_sqlite_ctgov2.R.. 50 tests OK 25.9s
# test_ctrdata_sqlite_ctis.R.... 87 tests OK 1.2s
# test_ctrdata_sqlite_euctr.R... 118 tests OK 47.7s
# test_ctrdata_sqlite_isrctn.R.. 38 tests OK 12.0s
# test_euctr_error_sample.R..... 8 tests OK 0.2s
# All ok, 674 results (4m 36.8s)
covr::package_coverage(path = ".", type = "tests")
# ctrdata Coverage: 94.27%
# R/ctrShowOneTrial.R: 57.89%
# R/dbGetFieldsIntoDf.R: 80.14%
# R/zzz.R: 80.95%
# R/ctrRerunQuery.R: 86.23%
# R/ctrGetQueryUrl.R: 89.32%
# R/util_functions.R: 89.84%
# R/ctrLoadQueryIntoDbEuctr.R: 90.08%
# R/ctrFindActiveSubstanceSynonyms.R: 90.38%
# R/ctrLoadQueryIntoDbCtgov2.R: 92.68%
# R/ctrLoadQueryIntoDbIsrctn.R: 92.81%
# R/ctrLoadQueryIntoDbCtis.R: 95.34%
# R/dbFindFields.R: 95.88%
# R/f_primaryEndpointResults.R: 96.00%
# R/dfMergeVariablesRelevel.R: 96.55%
# R/ctrLoadQueryIntoDb.R: 96.86%
# R/ctrGenerateQueries.R: 97.32%
# R/ctrOpenSearchPagesInBrowser.R: 97.40%
# R/dbFindIdsUniqueTrials.R: 98.78%
# R/f_numTestArmsSubstances.R: 98.92%
# R/f_likelyPlatformTrial.R: 99.13%
# R/dbQueryHistory.R: 100.00%
# R/dfName2Value.R: 100.00%
# R/dfTrials2Long.R: 100.00%
# R/f_controlType.R: 100.00%
# R/f_isMedIntervTrial.R: 100.00%
# R/f_isUniqueTrial.R: 100.00%
# R/f_numSites.R: 100.00%
# R/f_primaryEndpointDescription.R: 100.00%
# R/f_resultsDate.R: 100.00%
# R/f_sampleSize.R: 100.00%
# R/f_sponsorType.R: 100.00%
# R/f_startDate.R: 100.00%
# R/f_statusRecruitment.R: 100.00%
# R/f_trialObjectives.R: 100.00%
# R/f_trialPhase.R: 100.00%
# R/f_trialPopulation.R: 100.00%
# R/f_trialTitle.R: 100.00%
Future features
-
See project outline https://github.com/users/rfhb/projects/1
-
Authentication, expected to be required by CTGOV2; specifications not
yet known (work not yet started).
-
Explore further registers (exploration is continually ongoing; added
value, terms and conditions for programmatic access vary; no clear
roadmap is established yet).
Implemented:
-
~~Retrieve previous versions of protocol- or results-related
information. The challenges include, historic versions can only be
retrieved one-by-one, do not include results, or are not in structured
format. The functionality available at this time, is to retrieve any
version in CTGOV2, and to create a version when re-running a CTIS
query and retrieving new data.~~
-
~~Canonical definitions, filters, calculations are in the works (since
August 2023, released in version 1.20.0) for data mangling and
analyses across registers, e.g. to define study population, identify
interventional trials, calculate study duration; public collaboration
on these canonical scripts will speed up harmonising analyses.~~
-
~~Merge results-related fields retrieved from different registers,
such as corresponding endpoints. The challenge is the incomplete
congruency and different structure of data fields. This has largely
been implemented as per the previous bullet point.~~
Acknowledgements
-
Data providers and curators of the clinical trial registers. Please
review and respect their copyrights and terms and conditions, see
ctrOpenSearchPagesInBrowser(copyright = TRUE).
-
Package ctrdata has been made possible building on the work done for
R,
clipr.
curl,
dplyr,
duckdb,
httr,
jqr,
jsonlite,
lubridate,
mongolite,
nodbi,
RPostgres,
RSQLite,
rvest,
stringi and
xml2.
Issues and notes
-
Information in trial registers may not be fully correct; see for
example this publication on
CTGOV.
-
A warning may be issued and a record not imported if the complexity of
the XML content is too high for processing. The issue can be resolved
by increasing in the operating system the stack size available to R,
see: https://github.com/rfhb/ctrdata/issues/22
-
Please file issues and bugs
here. Also check out how to
handle some of the closed issues, e.g. on C stack usage too close to
the limit and on a SSL
certificate problem: unable to get local issuer
certificate.
Trial records in databases
SQLite
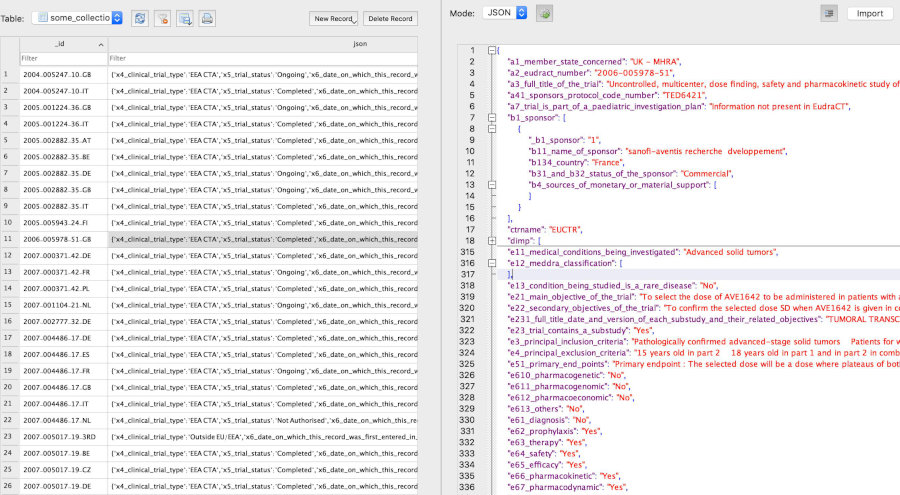 Example JSON representation in
SQLite
Example JSON representation in
SQLite
MongoDB
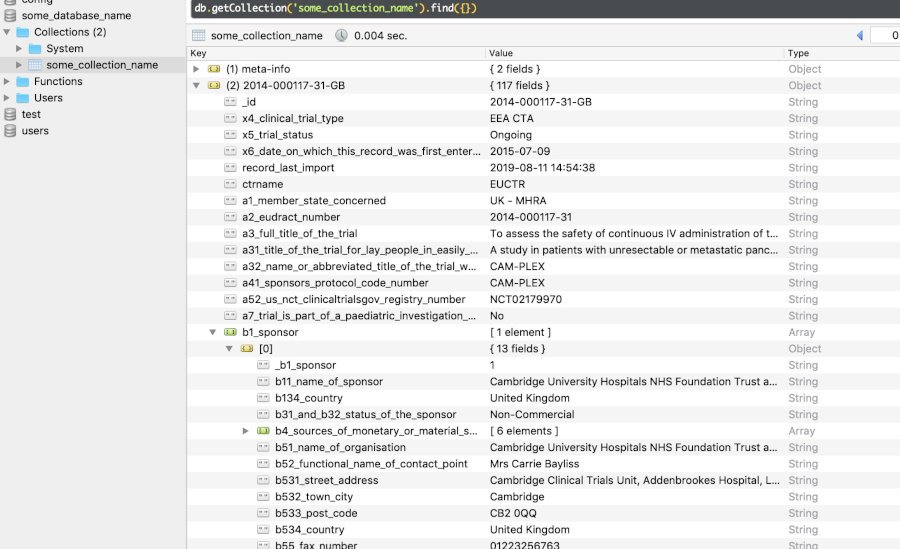 Example JSON representation in
MongoDB
Example JSON representation in
MongoDB
Try the ctrdata package in your browser
Any scripts or data that you put into this service are public.
ctrdata documentation built on June 8, 2025, 10:45 a.m.
Main features • References • Installation • Overview • Databases • Data model • Example workflow • Analysis across registers • Tests • Acknowledgements • Future
ctrdata for aggregating and analysing clinical trials
The package ctrdata provides functions for retrieving (downloading),
aggregating and analysing clinical trials using information (structured
protocol and result data, as well as documents) from public registers.
It can be used with the
- EU Clinical Trials Register (“EUCTR”, https://www.clinicaltrialsregister.eu/)
- EU Clinical Trials Information System (“CTIS”, https://euclinicaltrials.eu/, see example)
- ClinicalTrials.gov (“CTGOV2”, see example)
- ISRCTN Registry (https://www.isrctn.com/)
The motivation is to investigate the design and conduct of trials of
interest, to describe their trends and availability for patients and to
facilitate using their detailed results for research and meta-analyses.
ctrdata is a package for the R system,
but other systems and tools can use the databases created with this
package. This README was reviewed on 2025-06-07 for version 1.22.2.9000.
Main features
-
Protocol- and results-related trial information is easily downloaded: Users define a query in a register’s web interface, then copy the URL and enter it into
ctrdatawhich retrieves in one go all trials found. A script can automate copying the query URL from all registers. Personal annotations can be made when downloading trials. Also, trial documents and historic versions as available in registers on trials can be downloaded. -
Downloaded trial information is transformed and stored in a collection of a document-centric database, for fast and offline access. Information from different registers can be accumulated in a single collection. Uses
DuckDB,PostgreSQL,RSQLiteorMongoDB, via R packagenodbi: see section Databases below. Interactively browse through trial structure and data. Easily re-run any previous query in a collection to retrieve and update trial records. -
For analyses, convenience functions in
ctrdataimplement canonical trial concepts to simplify analyses across registers 🔔, allow find synonyms of an active substance, identify unique (de-duplicated) trial records across all registers, to merge and recode fields as well as to easily access deeply-nested fields. Analysis can be done withR(see vignette) or other systems, using theJSON-structured information in the database.
Remember to respect the registers’ terms and conditions (see
ctrOpenSearchPagesInBrowser(copyright = TRUE)). Please cite this
package in any publication as follows: “Ralf Herold (2025). ctrdata:
Retrieve and Analyze Clinical Trials in Public Registers. R package
version 1.22.2, https://cran.r-project.org/package=ctrdata”.
References
Package ctrdata has been used for unpublished works and these
publications:
- Jong et al. (2025) Experiences with Low-Intervention Clinical Trials—the New Category under the European Union Clinical Trials Regulation. Clinical Trials https://doi.org/10.1177/17407745241309293
- Lopez-Rey et al. (2025) Use of Bayesian Approaches in Oncology Clinical Trials: A Cross-Sectional Analysis’. Frontiers in Pharmacology https://doi.org/10.3389/fphar.2025.1548997
- Russek et al. (2025) Supplementing Single-Arm Trials with External Control Arms—Evaluation of German Real-World Data. Clinical Pharmacology & Therapeutics https://doi.org/10.1002/cpt.3684
- Alzheimer’s disease Horizon Scanning Report (2024) PDF file, p 10
- Kundu et al. (2024) Analysis of Factors Influencing Enrollment Success in Hematology Malignancy Cancer Clinical Trials (2008-2023). Blood Meeting Abstracts https://doi.org/10.1182/blood-2024-207446
- Lasch et al. (2022) The Impact of COVID‐19 on the Initiation of Clinical Trials in Europe and the United States. Clinical Pharmacology & Therapeutics https://doi.org/10.1002/cpt.2534
- Sood et al. (2022) Managing the Evidence Infodemic: Automation Approaches Used for Developing NICE COVID-19 Living Guidelines. medRxiv https://doi.org/10.1101/2022.06.13.22276242
- Blogging on Innovation coming to paediatric research
- Cancer Research UK (2017) The impact of collaboration: The value of UK medical research to EU science and health
- EMA (2017) Results of juvenile animal studies (JAS) and impact on anti-cancer medicine development and use in children PDF file, p 34
Installation
1. Install package ctrdata in R
Package ctrdata is on
CRAN and on
GitHub. Within
R, use the following commands to install
package ctrdata:
# Install CRAN version:
install.packages("ctrdata")
# Alternatively, install development version:
install.packages("devtools")
devtools::install_github("rfhb/ctrdata", build_vignettes = TRUE)
These commands also install the package’s dependencies (jsonlite,
httr, curl, clipr, xml2, nodbi, stringi, tibble,
lubridate, jqr, dplyr, zip, readr, digest, countrycode,
htmlwidgets, stringdist and V8).
2. Script to automatically copy user’s query from web browser
Optional; works with all registers supported by ctrdata and is
recommended for CTIS so that its URL in the web browser reflects the
user’s parameters for querying this register.
In the web browser, install the Tampermonkey browser
extension, click on “New user script”
and then on “Tools”, enter into “Import from URL” this URL:
https://raw.githubusercontent.com/rfhb/ctrdata/master/tools/ctrdataURLcopier.js
and then click on “Install”.
The browser extension can be disabled and enabled by the user. When
enabled, the URLs to all user’s queries in the registers are
automatically copied to the clipboard and can be pasted into the
queryterm = ... parameter of function
ctrLoadQueryIntoDb().
Additionally, this script retrieves results for CTIS when opening
search URLs such as
https://euclinicaltrials.eu/ctis-public/search#searchCriteria={"status":[3,4]}.
After changing the URL in the browser, a “Reload page” is needed to
conduct the search and show results.
Overview of functions in ctrdata
The functions are listed in the approximate order of use in a user’s workflow (in bold, main functions). See also the package documentation overview.
| Function name | Function purpose |
|----|----|
| ctrOpenSearchPagesInBrowser() | Open search pages of registers or execute search in web browser |
| ctrFindActiveSubstanceSynonyms() | Find synonyms and alternative names for an active substance |
| ctrGenerateQueries() | 🔔 From simple user parameters, generates queries for each register to find trials of interest |
| ctrGetQueryUrl() | Import from clipboard the URL of a search in one of the registers |
| ctrLoadQueryIntoDb() | Retrieve (download) or update, and annotate, information on trials from a register and store in a collection in a database |
| ctrShowOneTrial() | 🔔 Show full structure and all data of a trial, interactively select fields of interest for dbGetFieldsIntoDf() |
| dbQueryHistory() | Show the history of queries that were downloaded into the collection |
| dbFindIdsUniqueTrials() | Get the identifiers of de-duplicated trials in the collection |
| dbFindFields() | Find names of variables (fields) in the collection |
| dbGetFieldsIntoDf() | Create a data frame (or tibble) from trial records in the database with the specified fields |
| dfTrials2Long() | Transform the data.frame from dbGetFieldsIntoDf() into a long name-value data.frame, including deeply nested fields |
| dfName2Value() | From a long name-value data.frame, extract values for variables (fields) of interest (e.g., endpoints) |
| dfMergeVariablesRelevel() | Merge variables into a new variable, optionally map values to a new set of levels |
Databases for use with ctrdata
Package ctrdata retrieves trial information and stores it in a
database collection, which has to be given as a connection object to
parameter con for several ctrdata functions. This connection object
is created almost identically for the four database backends supported
by ctrdata, as shown in the table. For a speed comparison, see the
nodbi documentation.
Besides ctrdata functions below, such a connection object can be used
with functions of other packages, such as nodbi (see last row in
table) or, in case of MongoDB as database backend, mongolite (see
vignettes).
| Purpose | Function call |
|----|----|
| Create SQLite database connection | dbc <- nodbi::src_sqlite(dbname = "name_of_my_database", collection = "name_of_my_collection") |
| Create DuckDB database connection | dbc <- nodbi::src_duckdb(dbdir = "name_of_my_database", collection = "name_of_my_collection") |
| Create MongoDB database connection | dbc <- nodbi::src_mongo(db = "name_of_my_database", collection = "name_of_my_collection") |
| Create PostgreSQL database connection | dbc <- nodbi::src_postgres(dbname = "name_of_my_database"); dbc[["collection"]] <- "name_of_my_collection" |
| Use connection with ctrdata functions | ctrdata::{ctrLoadQueryIntoDb, dbQueryHistory, dbFindIdsUniqueTrials, dbFindFields, dbGetFieldsIntoDf}(con = dbc, ...) |
| Use connection with nodbi functions | e.g., nodbi::docdb_query(src = dbc, key = dbc$collection, ...) |
Data model of ctrdata
Package ctrdata uses the data models that are implicit in data as
retrieved from the different registers. No mapping is provided for any
register’s data model to a putative target data model. The reasons
include that registers’ data models are continually evolving over time,
that only few data fields have similar values and meaning between
registers, and that the retrieved public data may not correspond to the
registers’ internal data model. The structure of data for a specific
trial can interactively be inspected and searched using function, see
the section below.
Thus, the handling of data from different models of registers is to be
done at the time of analysis. This approach allows a high level of
flexibility, transparency and reproducibility. To support analyses,
ctrdata (from version 1.21.0) provides functions that calculate
concepts of clinical trials across registers, which are commonly used in
analyses, such as start dates, age groups and statistical tests of
results. See
help(ctrdata-trial-concepts)
and the section Analysis across
trials in the example workflow
below. For further analyses, see examples of function
dfMergeVariablesRelevel()
on how to align related fields from different registers for a joint
analysis.
In any of the
databases,
one clinical trial is one document, corresponding to one row in a
SQLite, PostgreSQL or DuckDB table, and to one document in a
MongoDB collection. These NoSQL backends allow documents to have
different structures, which is used here to accommodate the different
models of data retrieved from the registers. Package ctrdata stores in
every such document:
- field
_idwith the trial identification as provided by the register from which it was retrieved - field
ctrnamewith the name of the register (EUCTR,CTGOV,CTGOV2,ISRCTN,CTIS) from which that trial was retrieved - field
record_last_importwith the date and time when that document was last updated usingctrLoadQueryIntoDb() - only for
CTGOV2andCTIS: objecthistorywith a historic version of the trial and withhistory_version, which contains the fieldsversion_number(starting from 1) andversion_date - all original fields as provided by the register for that trial (see example in vignette)
For visualising the data structure for a trial, see this vignette section.
Vignettes
- Install R package ctrdata
- Retrieve clinical trial information
- Summarise and analyse clinical trial information
Example workflow
The aim is to download protocol-related trial information and tabulate the trials’ status of conduct.
- Attach package
ctrdata:
library(ctrdata)
- See help to get started with
ctrdata:
help("ctrdata")
- Information on trial registers, their contents and how they can be
used with
ctrdata(last updated 2025-05-31):
help("ctrdata-registers")
- Trial concepts across registers provided by
ctrdata(new 2025-03-09 🔔):
help("ctrdata-trial-concepts")
- Open registers’ advanced search pages in browser:
ctrOpenSearchPagesInBrowser()
# Please review and respect register copyrights:
ctrOpenSearchPagesInBrowser(copyright = TRUE)
-
Adjust search parameters and execute search in browser
-
When trials of interest are listed in browser, copy the address from the browser’s address bar to the clipboard (you can automate this, see here)
-
Search used in this example: https://www.clinicaltrialsregister.eu/ctr-search/search?query=neuroblastoma&phase=phase-two&age=children
-
Get address from clipboard:
q <- ctrGetQueryUrl()
# * Using clipboard content as register query URL: https://www.clinicaltrialsregister.eu/
# ctr-search/search?query=neuroblastoma&phase=phase-two&age=children
# * Found search query from EUCTR: query=neuroblastoma&phase=phase-two&age=children
q
# query-term query-register
# 1 query=neuroblastoma&phase=phase-two&age=children EUCTR
🔔 Queries in the trial registers can automatically copied to the clipboard (including for “CTIS”, where the URL otherwise does not show the user’s query) using the solution here.
- Retrieve protocol-related information, transform and save to database:
For loading the trial information, first a database collection is
specified, using nodbi (see above for how to specify PostgreSQL,
RSQlite, DuckDB or MongoDB as backend, see section
Databases):
# Connect to (or create) an SQLite database
# stored in a file on the local system:
db <- nodbi::src_sqlite(
dbname = "database_name.sql",
collection = "collection_name"
)
Then, the trial information is retrieved and loaded into the collection:
# Retrieve trials from public register:
ctrLoadQueryIntoDb(
queryterm = q,
euctrresults = TRUE,
con = db
)
# * Checking trials in EUCTR...
# Retrieved overview, multiple records of 73 trial(s) from 4 page(s) to be downloaded (estimate: 9 MB)
# (1/3) Downloading trials...
# Note: register server cannot compress data, transfer takes longer (estimate: 90 s)
# Download status: 4 done; 0 in progress. Total size: 6.39 Mb (100%)... done!
# (2/3) Converting to NDJSON (estimate: 1 s)...
# (3/3) Importing records into database...
# = Imported or updated 270 records on 73 trial(s)
# * Checking results if available from EUCTR for 73 trials:
# (1/4) Downloading results...
# Download status: 73 done; 0 in progress. Total size: 22.57 Mb (100%)... done!
# Download status: 41 done; 0 in progress. Total size: 165.04 Kb (100%)... done!
# Download status: 41 done; 0 in progress. Total size: 165.04 Kb (100%)... done!
# - extracting results (. = data, F = file[s] and data, x = none):
# F F . . . . . . . F . . . F . . . F . . . . . F . . . . . . . F
# (2/4) Converting to NDJSON (estimate: 3 s)...
# (3/4) Importing results into database (may take some time)...
# (4/4) Results history: not retrieved (euctrresultshistory = FALSE)
# = Imported or updated results for 32 trials
# No history found in expected format.
# Updated history ("meta-info" in "some_collection_name")
# $n
# [1] 270
Under the hood, EUCTR plain text and XML files from EUCTR, CTGOV, ISRCTN
are converted using Javascript via V8 in R into NDJSON, which is
imported into the database collection.
- Easily generate queries for each register and add records from several registers into the same collection
The same parameters can be used to ask ctrdata to generate search
queries that apply to each register, for opening the web interfaces and
for loading the trial data into the collection:
# Generate queries for each register
queries <- ctrGenerateQueries(
condition = "neuroblastoma",
recruitment = "completed",
phase = "phase 2",
population = "P"
)
queries
# EUCTR
# "https://www.clinicaltrialsregister.eu/ctr-search/search?query=neuroblastoma&phase=phase-two&age=children&age=adolescent&age=infant-and-toddler&age=newborn&age=preterm-new-born-infants&age=under-18&status=completed"
# ISRCTN
# "https://www.isrctn.com/search?&q=&filters=condition:neuroblastoma,phase:Phase II,ageRange:Child,trialStatus:completed,primaryStudyDesign:Interventional"
# CTGOV2
# "https://clinicaltrials.gov/search?cond=neuroblastoma&aggFilters=phase:2,ages:child,status:com,studyType:int"
# CTGOV2expert
# "https://clinicaltrials.gov/expert-search?term=AREA[ConditionSearch]\"neuroblastoma\" AND (AREA[Phase]\"PHASE2\") AND (AREA[StdAge]\"CHILD\") AND (AREA[OverallStatus]\"COMPLETED\") AND (AREA[StudyType]INTERVENTIONAL)"
# CTIS
# "https://euclinicaltrials.eu/ctis-public/search#searchCriteria={\"medicalCondition\":\"neuroblastoma\",\"trialPhaseCode\":[4],\"ageGroupCode\":[2],\"status\":[5,8]}"
# Open queries in registers' web interfaces
sapply(queries, ctrOpenSearchPagesInBrowser)
# Load all queries into database collection
result <- lapply(queries, ctrLoadQueryIntoDb, con = db)
sapply(result, "[[", "n")
# EUCTR ISRCTN CTGOV2 CTGOV2expert CTIS
# 180 0 110 110 1
- Analyse
Tabulate the status of trials that are part of an agreed paediatric
development program (paediatric investigation plan, PIP). ctrdata
functions return a data.frame (or a tibble, if package tibble is
loaded).
# Get all records that have values in the fields of interest:
result <- dbGetFieldsIntoDf(
# Field of interest
fields = c("a7_trial_is_part_of_a_paediatric_investigation_plan"),
# Trial concepts calculated across registers
calculate = c("f.statusRecruitment", "f.isUniqueTrial"),
con = db
)
# Querying database (16 fields)...
# Searching for duplicate trials...
# - Getting all trial identifiers (may take some time), 381 found in collection
# - Finding duplicates among registers' and sponsor ids...
# - 174 EUCTR _id were not preferred EU Member State record for 66 trials
# - Keeping 110 / 60 / 0 / 0 / 0 records from CTGOV2 / EUCTR / CTGOV / ISRCTN / CTIS
# = Returning keys (_id) of 170 records in collection "collection_name"
# Tabulate the clinical trial information of interest
with(
result[result$.isUniqueTrial, ],
table(
.statusRecruitment,
a7_trial_is_part_of_a_paediatric_investigation_plan
)
)
# a7_trial_is_part_of_a_paediatric_investigation_plan
# .statusRecruitment FALSE TRUE
# ongoing 2 3
# completed 12 5
# ended early 7 3
# other 9 3
- Queries to CTGOV and CTGOV2
The new website and API introduced in July 2023
(https://www.clinicaltrials.gov/) is supported by ctrdata since
mid-2023 and identified in ctrdata as CTGOV2.
On 2024-06-25, CTGOV has retired the classic website and API used by
ctrdata since 2015. To support users, ctrdata automatically
translates and redirects queries to the current website. This helps with
automatically updating previously loaded queries
(ctrLoadQueryIntoDb(querytoupdate = <n>)), manually migrating queries
and reproducible work on clinical trials information. Going forward,
users are recommended to change to use CTGOV2 queries.
As regards study data, important differences exist between field names
and contents of information retrieved using CTGOV or CTGOV2; see the
schema for study protocols in
CTGOV,
the schema for study
results
and the Study Data Structure for
CTGOV2.
For more details, call help("ctrdata-registers"). This is one of the
reasons why ctrdata handles the situation as if these were two
different registers and will continue to identify the current API as
register = "CTGOV2", to support the analysis stage.
Note that loading trials with ctrdata overwrites the previous record
with CTGOV2 data, whether the previous record was retrieved using
CTGOV or CTGOV2 queries.
Example using a CTGOV query:
# CTGOV search query URL
q <- "https://classic.clinicaltrials.gov/ct2/results?cond=neuroblastoma&rslt=With&recrs=e&age=0&intr=Drug"
# Open old URL (CTGOV) in current website (CTGOV2):
ctrOpenSearchPagesInBrowser(q)
# * Appears specific for CTGOV Classic website
# Since 2024-06-25, the classic CTGOV servers are no longer available.
# Package ctrdata has translated the classic CTGOV query URL from this call
# of function ctrLoadQueryIntoDb(queryterm = ...) into a query URL that works
# with the current CTGOV2. This is printed below and is also part of the return
# value of this function, ctrLoadQueryIntoDb(...)$url. This URL can be used with
# ctrdata functions. Note that the fields and data schema of trials differ
# between CTGOV and CTGOV2.
#
# Replace this URL:
#
# https://classic.clinicaltrials.gov/ct2/results?cond=neuroblastoma&rslt=With&recrs=e&age=0&intr=Drug
#
# with this URL:
#
# https://clinicaltrials.gov/search?cond=neuroblastoma&intr=Drug&aggFilters=ages:child,results:with,status:com
#
# * Found search query from CTGOV2: cond=neuroblastoma&intr=Drug&aggFilters=ages:child,results:with,status:com
# Count trials:
ctrLoadQueryIntoDb(
queryterm = q,
con = db,
only.count = TRUE
)
# $n
# [1] 71
- Queries to CTIS
Queries in the CTIS search interface can be automatically copied to the
clipboard so that a user can paste them into queryterm, see
here.
Subsequent to the relaunch of CTIS on 2024-07-18, there are more than
8,700 trials publicly accessible in CTIS. See
below for how to download documents from CTIS.
# See how many trials are in CTIS publicly accessible:
ctrLoadQueryIntoDb(
queryterm = "",
register = "CTIS",
only.count = TRUE
)
# $n
# [1] 9181
- Inspect and search structure of trial information
For a given trial, function ctrShowOneTrial() enables the user to visualise the hiearchy of fields and contents in the user’s local web browser, to search for field names and field values, and to select and copy selected fields’ names for use with function dbGetFieldsIntoDf().
# This opens a local browser for user interaction.
# If the trial identifier (_id) is not found in the
# specified collection, it will be retrieved from the register.
ctrShowOneTrial(
identifier = "2024-518931-12-00",
con = db
)
- Analysis across registers
Show cumulative start of trials over time. This uses the calculation of
trial concepts as available since ctrdata version 1.21.0 🔔.
# use helper package
library(dplyr)
library(ggplot2)
df <- dbGetFieldsIntoDf(
fields = "",
calculate = c("f.statusRecruitment", "f.isUniqueTrial", "f.startDate"),
con = db
)
df %>%
filter(.isUniqueTrial) %>%
ggplot() +
stat_ecdf(aes(
x = .startDate,
colour = .statusRecruitment
)) +
labs(
title = "Evolution over time of selected trials",
subtitle = "Data from EUCTR, CTIS, ISRCTN, CTGOV2",
x = "Date of start (proposed or realised)",
y = "Cumulative proportion of trials",
colour = "Current status",
caption = Sys.Date()
)
ggsave(
filename = "man/figures/README-ctrdata_across_registers.png",
width = 5, height = 3, units = "in"
)
 Analysis across registers
Analysis across registers
- Result-related trial information
Analyse some simple result details, here from CTGOV2 (see this vignette for more examples):
# use helper package
library(ggplot2)
result <- dbGetFieldsIntoDf(
calculate = c(
"f.numSites",
"f.sampleSize",
"f.controlType",
"f.numTestArmsSubstances"
),
con = db
)
ggplot(data = result) +
labs(
title = "Selected trials",
subtitle = "Patients with a neuroblastoma"
) +
geom_point(
mapping = aes(
x = .numSites,
y = .sampleSize,
size = .numTestArmsSubstances,
colour = .controlType
)
) +
scale_x_log10() +
scale_y_log10() +
labs(
x = "Number of sites",
y = "Total number of participants",
colour = "Control",
size = "# Treatments",
caption = Sys.Date()
)
ggsave(
filename = "man/figures/README-ctrdata_results_neuroblastoma.png",
width = 5, height = 3, units = "in"
)
 Neuroblastoma trials
Neuroblastoma trials
- Download documents: retrieve protocols, statistical analysis plans and
other documents into the local folder
./files-.../
### EUCTR document files can be downloaded when results are requested
# All files are downloaded and saved (documents.regexp is not used with EUCTR)
ctrLoadQueryIntoDb(
queryterm = "query=cancer&age=under-18&phase=phase-one",
register = "EUCTR",
euctrresults = TRUE,
documents.path = "./files-euctr/",
con = db
)
# * Found search query from EUCTR: query=cancer&age=under-18&phase=phase-one
# * Checking trials in EUCTR...
# [...]
# = documents saved in './files-euctr'
# Updated history ("meta-info" in "some_collection_name")
### CTGOV files are downloaded, here corresponding to the default of
# documents.regexp = "prot|sample|statist|sap_|p1ar|p2ars|ctalett|lay|^[0-9]+ "
ctrLoadQueryIntoDb(
queryterm = "cond=Neuroblastoma&type=Intr&recrs=e&phase=1&u_prot=Y&u_sap=Y&u_icf=Y",
register = "CTGOV",
documents.path = "./files-ctgov/",
con = db
)
# * Checking for documents...
# - Getting links to documents
# - Downloading documents into 'documents.path' = ./files-ctgov/
# - Created directory ./files-ctgov
# - Applying 'documents.regexp' to 40 missing documents
# - Creating subfolder for each trial
# - Downloading 40 missing documents
# Download status: 40 done; 0 in progress. Total size: 110.75 Mb (100%)... done!
# = Newly saved 40 document(s) for 32 trial(s); 0 of such document(s) for 0 trial(s)
# already existed in ./files-ctgov
### CTGOV2 files are downloaded, using the default of documents.regexp
ctrLoadQueryIntoDb(
queryterm = "https://clinicaltrials.gov/search?cond=neuroblastoma&aggFilters=phase:1,results:with",
documents.path = "./files-ctgov2/",
con = db
)
# * Checking for documents...
# - Getting links to documents
# - Downloading documents into 'documents.path' = ./files-ctgov2/
# - Created directory ./files-ctgov2
# - Creating subfolder for each trial
# - Applying 'documents.regexp' to 42 missing documents
# - Downloading 42 missing documents
# Download status: 42 done; 0 in progress. Total size: 92.57 Mb (100%)... done!
# = Newly saved 42 document(s) for 26 trial(s); 0 of such document(s) for 0
# trial(s) already existed in ./files-ctgov2
### ISRCTN files are downloaded, using the default of documents.regexp
ctrLoadQueryIntoDb(
queryterm = "https://www.isrctn.com/search?q=alzheimer",
documents.path = "./files-isrctn/",
con = db
)
# * Found search query from ISRCTN: q=alzheimer
# [...]
# * Checking for documents...
# - Getting links to documents
# - Downloading documents into 'documents.path' = ./files-isrctn/
# - Created directory /Users/ralfherold/Daten/mak/r/emea/ctrdata/files-isrctn
# - Applying 'documents.regexp' to 52 missing documents
# - Creating subfolder for each trial
# - Downloading 32 missing documents
# Download status: 32 done; 0 in progress. Total size: 14.89 Mb (100%)... done!
# = Newly saved 26 document(s) for 15 trial(s); 0 of such document(s) for 0
# trial(s) already existed in ./files-isrctn
### CTIS files are downloaded, using the default of documents.regexp
ctrLoadQueryIntoDb(
queryterm = paste0(
"https://euclinicaltrials.eu/ctis-public/search#",
'searchCriteria={"containAny":"cancer","status":[8]}'
),
documents.path = "./files-ctis/",
documents.regexp = "icf",
con = db
)
# * Found search query from CTIS: searchCriteria={"containAny":"cancer"}
# [...]
# * Checking for documents: . . . . . . . . . . . . . . . . . . .
# - Downloading documents into 'documents.path' = ./files-ctis/
# - Applying 'documents.regexp' to 1114 missing documents
# - Creating subfolder for each trial
# - Downloading 512 missing documents (excluding 2 files with duplicate names
# for saving, e.g. /Users/ralfherold/Daten/mak/r/emea/ctrdata/files-ctis/2022-
# 500694-14-00/SbjctInfaICF - L1 SIS and ICF Prescreening ICF clean placeholder
# - 137297.PDF, /Users/ralfherold/Daten/mak/r/emea/ctrdata/files-ctis/2022-
# 500694-14-00/SbjctInfaICF - L1 SIS and ICF Pregnant Partner ICF clean -
# 137297.PDF)
# Download status: 510 done; 0 in progress. Total size: 377.27 Kb (100%)... done!
# Redirecting to CDN...
# Download status: 127 done; 0 in progress. Total size: 47.66 Mb (100%)... done!
# = Newly saved 510 document(s) for 35 trial(s); 0 of such document(s) for 0
# trial(s) already existed in ./files-ctis
Tests and coverage
See also https://app.codecov.io/gh/rfhb/ctrdata/tree/master/R
# 2025-05-17
tinytest::test_all()
# test_ctrdata_duckdb_ctgov2.R.. 78 tests OK 46.4s
# test_ctrdata_function_activesubstance.R 4 tests OK 0.8s
# test_ctrdata_function_ctrgeneratequeries.R 14 tests OK 18ms
# test_ctrdata_function_params.R 25 tests OK 0.8s
# test_ctrdata_function_trial-concepts.R 80 tests OK 3.2s
# test_ctrdata_function_various.R 76 tests OK 3.7s
# test_ctrdata_postgres_ctgov2.R 50 tests OK 32.4s
# test_ctrdata_sqlite_ctgov.R... 46 tests OK 29.0s
# test_ctrdata_sqlite_ctgov2.R.. 50 tests OK 25.9s
# test_ctrdata_sqlite_ctis.R.... 87 tests OK 1.2s
# test_ctrdata_sqlite_euctr.R... 118 tests OK 47.7s
# test_ctrdata_sqlite_isrctn.R.. 38 tests OK 12.0s
# test_euctr_error_sample.R..... 8 tests OK 0.2s
# All ok, 674 results (4m 36.8s)
covr::package_coverage(path = ".", type = "tests")
# ctrdata Coverage: 94.27%
# R/ctrShowOneTrial.R: 57.89%
# R/dbGetFieldsIntoDf.R: 80.14%
# R/zzz.R: 80.95%
# R/ctrRerunQuery.R: 86.23%
# R/ctrGetQueryUrl.R: 89.32%
# R/util_functions.R: 89.84%
# R/ctrLoadQueryIntoDbEuctr.R: 90.08%
# R/ctrFindActiveSubstanceSynonyms.R: 90.38%
# R/ctrLoadQueryIntoDbCtgov2.R: 92.68%
# R/ctrLoadQueryIntoDbIsrctn.R: 92.81%
# R/ctrLoadQueryIntoDbCtis.R: 95.34%
# R/dbFindFields.R: 95.88%
# R/f_primaryEndpointResults.R: 96.00%
# R/dfMergeVariablesRelevel.R: 96.55%
# R/ctrLoadQueryIntoDb.R: 96.86%
# R/ctrGenerateQueries.R: 97.32%
# R/ctrOpenSearchPagesInBrowser.R: 97.40%
# R/dbFindIdsUniqueTrials.R: 98.78%
# R/f_numTestArmsSubstances.R: 98.92%
# R/f_likelyPlatformTrial.R: 99.13%
# R/dbQueryHistory.R: 100.00%
# R/dfName2Value.R: 100.00%
# R/dfTrials2Long.R: 100.00%
# R/f_controlType.R: 100.00%
# R/f_isMedIntervTrial.R: 100.00%
# R/f_isUniqueTrial.R: 100.00%
# R/f_numSites.R: 100.00%
# R/f_primaryEndpointDescription.R: 100.00%
# R/f_resultsDate.R: 100.00%
# R/f_sampleSize.R: 100.00%
# R/f_sponsorType.R: 100.00%
# R/f_startDate.R: 100.00%
# R/f_statusRecruitment.R: 100.00%
# R/f_trialObjectives.R: 100.00%
# R/f_trialPhase.R: 100.00%
# R/f_trialPopulation.R: 100.00%
# R/f_trialTitle.R: 100.00%
Future features
-
See project outline https://github.com/users/rfhb/projects/1
-
Authentication, expected to be required by CTGOV2; specifications not yet known (work not yet started).
-
Explore further registers (exploration is continually ongoing; added value, terms and conditions for programmatic access vary; no clear roadmap is established yet).
Implemented:
-
~~Retrieve previous versions of protocol- or results-related information. The challenges include, historic versions can only be retrieved one-by-one, do not include results, or are not in structured format. The functionality available at this time, is to retrieve any version in CTGOV2, and to create a version when re-running a CTIS query and retrieving new data.~~
-
~~Canonical definitions, filters, calculations are in the works (since August 2023, released in version 1.20.0) for data mangling and analyses across registers, e.g. to define study population, identify interventional trials, calculate study duration; public collaboration on these canonical scripts will speed up harmonising analyses.~~
-
~~Merge results-related fields retrieved from different registers, such as corresponding endpoints. The challenge is the incomplete congruency and different structure of data fields. This has largely been implemented as per the previous bullet point.~~
Acknowledgements
-
Data providers and curators of the clinical trial registers. Please review and respect their copyrights and terms and conditions, see
ctrOpenSearchPagesInBrowser(copyright = TRUE). -
Package
ctrdatahas been made possible building on the work done for R, clipr. curl, dplyr, duckdb, httr, jqr, jsonlite, lubridate, mongolite, nodbi, RPostgres, RSQLite, rvest, stringi and xml2.
Issues and notes
-
Information in trial registers may not be fully correct; see for example this publication on CTGOV.
-
A warning may be issued and a record not imported if the complexity of the XML content is too high for processing. The issue can be resolved by increasing in the operating system the stack size available to R, see: https://github.com/rfhb/ctrdata/issues/22
-
Please file issues and bugs here. Also check out how to handle some of the closed issues, e.g. on C stack usage too close to the limit and on a SSL certificate problem: unable to get local issuer certificate.
Trial records in databases
SQLite
 Example JSON representation in
SQLite
Example JSON representation in
SQLite
MongoDB
 Example JSON representation in
MongoDB
Example JSON representation in
MongoDB
Try the ctrdata package in your browser
Any scripts or data that you put into this service are public.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
