LAScatalog formal class
In lidR: Airborne LiDAR Data Manipulation and Visualization for Forestry Applications
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
dev.args = list(pointsize = 10)
)
options(rmarkdown.html_vignette.check_title = FALSE)
library(lidR)
data = data.frame(
Max.X = c(885228.88, 886993.96, 885260.93, 887025.96,
885292.94, 887056.88, 892199.94, 893265.54, 892229.99, 893295.15,
888759.96, 890524.95, 892259.98, 894025.98, 892289.96, 894055.93,
888790.91, 890554.98, 888820.95, 890585.99, 892319.96, 894084.97,
892349.89, 894114.29, 895250.23, 895094.78, 895044.96, 895053.55,
885323.96, 887087.95, 885355.95, 887119.96, 883657.85, 885387.95,
887150.97, 885419.98, 887182.95, 883688.44, 885442.91, 887193.9,
888851.96, 890615.97, 888882.94, 890646.97, 892379.94, 894127.84,
892409.97, 892676.58, 888913.92, 890676.93, 888944.86, 890707.98,
892439.95, 894124.59, 892469.86, 894232.94, 894786.68, 888958.83,
890713.51, 892476.43, 894239.97, 894786.07),
Min.X = c(885022.37,
885204.73, 885027.52, 885229.03, 885040.86, 885261.03, 891503.09,
892198.69, 891501.42, 892200.07, 886970.07, 888735.55, 891499.96,
892230.05, 890521.99, 892260.01, 886994.05, 888760.09, 887026.07,
888791.01, 890525.05, 892290.04, 890555.01, 892320.12, 894002.98,
894026.02, 894056.02, 894085.03, 885051.45, 885293.03, 885063.29,
885324.03, 883166.09, 885072.16, 885356.09, 883642.36, 885388.15,
883180.23, 883658.11, 885420.02, 887057.07, 888821.02, 887088.11,
888852.03, 890586.03, 892350.02, 890616.07, 892380.01, 887120.07,
888883.03, 887151.11, 888914.02, 890647.06, 892410.06, 890677.07,
892440.07, 894209.19, 887183.07, 888945.12, 890708.03, 892470.16,
894233.07),
Max.Y = c(630219.48, 630214.96, 631609.95, 631604.97,
633001.65, 632995.99, 625898.35, 625882.94, 627289.82, 627273.89,
630174.88, 630134.94, 628681.66, 628664.99, 630094.95, 630057.95,
631564.98, 631524.94, 632955.82, 632915.99, 631486.9, 631447.96,
632876.93, 632838.96, 628627.89, 630019.93, 631410.97, 631740.88,
634393.05, 634386.96, 635786.24, 635779.96, 638613.36, 637176.84,
637169.92, 638601.99, 638560.96, 639938.36, 639926.95, 639558.31,
634346.93, 634307.92, 635739.92, 635699.92, 634268.97, 634229.95,
635659.89, 635622.88, 637129.84, 637089.93, 638520.94, 638481.91,
637051.99, 637012.92, 638442.98, 638403.94, 638366.87, 639177.04,
639133.74, 638702.56, 638702.56, 638702.56),
Min.Y = c(629157.18,
629099.31, 630215.04, 630175.05, 631605.02, 631565.05, 625816.52,
625793.6, 625883.01, 625860.81, 629036.82, 629017.72, 627274.01,
627251.36, 628665.04, 628628.01, 630135.08, 630095.02, 631525.01,
631487.19, 630058.02, 630020.05, 631448.08, 631411.03, 627506.32,
628612.41, 629999.84, 631390.38, 632996.06, 632956.04, 634387.01,
634347.01, 637939.24, 635780.07, 635740.05, 637170.11, 637130.14,
638602.13, 638561.04, 638521.07, 632916.05, 632877.04, 634308.06,
634269.04, 632839.06, 632801.04, 634230.04, 634223.9, 635700.07,
635660.11, 637090.03, 637052.15, 635623.06, 635619.13, 637013.1,
636979.71, 637259.33, 638482.01, 638443.02, 638404.08, 638367.11,
638355.37),
Min.Z = c(325.12, 251.48, 244.68, 286.7, 338.86, 320.68, 118.08, 60.69,
-5.01, -7.58, 225.29, 252, 87.3, 41.7, 115.01, 28.77, 205.11,
200.85, 200.54, 169.5, 90.64, 19.72, 126.04, 28.6, 41.98, 43.15,
7.74, 6.7, 199.26, 190.02, 284.92, 216.16, 218.14, 318.93, 220.21,
218.04, 137.31, 218.13, 217.34, 190.42, 207.62, 113.84, 118.18,
141.52, 92, 52.5, 91.2, 77.92, 156.57, 89.53, 108.83, 93.98,
23.45, 1.64, 33.22, 3.29, 0.61, 108.03, 208.38, 121.18, 58.83,
0.95),
Max.Z = c(418.46, 990.54, 409.06, 1021.87, 996.42, 1005.02, 173.77, 393.97,
836.52, 820.98, 414.2, 936.47, 792.95, 822.51, 777.31, 837.87,
419.15, 741.84, 907.2, 872.27, 898.53, 822.53, 846.77, 740.65,
826.61, 890.21, 828.86, 680.32, 390.31, 997.2, 965.55, 969.24,
249.34, 849.5, 950.2, 848.64, 904.1, 880, 827.92, 888.34, 462.88,
906.61, 440.83, 887.34, 860.37, 747.1, 808.75, 194.76, 734.21,
838.34, 834.76, 758.91, 771.76, 670.1, 810.94, 761.53, 109.26,
303.94, 349.94, 799.8, 737.01, 593.91),
Version.Major = 1,
Version.Minor = 2,
Point.Data.Format.ID = 1,
filename = paste0("path/to/las/files/file", 1:62, ".las")
)
geom <- lapply(1:nrow(data), function(i)
{
mtx <- matrix(c(data$Min.X[i], data$Max.X[i], data$Min.Y[i], data$Max.Y[i])[c(1, 1, 2, 2, 1, 3, 4, 4, 3, 3)], ncol = 2)
sf::st_polygon(list(mtx))
})
geom <-sf::st_sfc(geom)
sf::st_crs(geom) <- 26917
data <- sf::st_set_geometry(data, geom)
ctg <- new("LAScatalog")
ctg@data <- data
A LAScatalog class is a representation in R of a las file or a collection of las files not loaded in memory. Indeed, a regular computer cannot load the entire point cloud in R if it covers a broad area. For very high density datasets it can even fail loading a single file (see also the "LAS formal class" vignette). In lidR, we use a LAScatalog to process datasets that cannot fit in memory.
Build a LAScatalog object reading a folder of las files
ctg <- readLAScatalog("path/to/las/files/")
# or
ctg <- readLAScatalog("path/to/las/files/big_file.las")
ctg
Basic structure of a LAScatalog object
A LAScatalog contains a sf object with POLYGON geometries plus some extra slots that store information relative to how the LAScatalog will be processed.
The slot data of a LAScatalog object contains the sf object with the most important information read from the header of .las or .laz files. Reading only the header of the file provides an overview of the content of the files very quickly without actually loading the point cloud. The columns of the table are named after the LAS specification version 1.4
The other slots are well documented in the documentation help("LAScatalog-class") so they are not described in this vignette.
Allowed and non-allowed manipulation of a LAScatalog object
A LAScatalog purpose is not to manipulate spatial data in R. The purpose of a LAScatalog is to represent a set of existing las/laz files. Thus a LAScatalog cannot be modified because it must be related to the actual content of the files. The following throws an error:
ctg$Min.Z <- 0
Obviously it is always possible to modify an R object by bypassing such simple restrictions. In this case the user will break something internally and a correct output is not guaranteed.
However it is possible to add and modify the attributes using a name that is not reserved. The following is allowed:
ctg$newattribute <- 0
Validation of LAScatalog object
Users commonly report bugs arising from the fact that the point cloud is invalid. This is why we introduced the function las_check() to perform an inspection of the LAScatalog objects. This function checks if a LAScatalog object is consistent (files are all of the same type for example). For example, it may happen that a collection mixes files of type 1 with files of type 3 or files with different scale factors.
las_check(ctg)
The function las_check() when applied to a LAScatalog does not perform a deep inspection of the point cloud unlike when applied to a LAS object. Indeed the point cloud is not actually read.
Display a LAScatalog object
lidR provides a simple plot() function to plot a LAScatalog object:
plot(ctg)
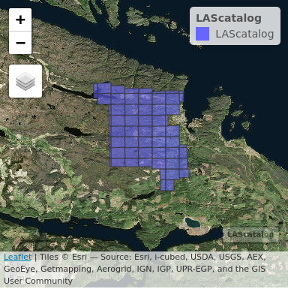
The option mapview = TRUE displays the LAScatalog on an interactive map with pan and zoom and allows the addition of a satellite map in the background. It uses the package mapview internally. It is often useful to check if the CRS of the file are properly registered. The epsg codes recorded in the las files appear to be sometime incorrect, according to our own experience.
plot(ctg, mapview = TRUE, map.type = "Esri.WorldImagery")
Using a sf object to store the attributes of the las file it is easy to display metadata of the files. In the following we can immediately see that the catalog is not normalized and is likely to contain outliers:
plot(ctg["Min.Z"])
Apply lidR functions on a LAScatalog
Most of lidR functions are compatible with a LAScatalog and work almost like with a single point cloud loaded in memory. In the following example we use the function pixel_metrics() to compute the mean elevation of the points. The output is a continuous wall-to-wall raster. It works exactly as if the input was a LAS object.
hmean <- pixel_metrics(ctg, mean(Z), 20)
However, processing a LAScatalog usually requires some tuning of the processing options to get better control of the computation. Indeed, if the catalog is huge the output is likely to be huge as well, and maybe the output cannot fit in the R memory. For example, normalize_height() throws an error if used 'as is' without tuning the processing options. Using normalize_height() like in the following example the expected output would be a huge point cloud loaded in memory. The lidR package forbids such a call:
output <- normalize_height(ctg, tin())
Instead, one can use the processing option opt_output_files(). Processing options drive how the big files are split in small chunks and how the outputs are either returned into R or written on disk into files.
opt_output_files(ctg) <- "folder/where/to/store/outputs/{ORIGINALFILENAME}_normalized"
output <- normalize_height(ctg, tin())
Here the output is not a point cloud but a LAScatalog pointing to the newly created files. The user can check how the collection will be processed by calling summary
summary(ctg)
Also the plot function can displays the chunks pattern i.e. how the dataset is split into small chunks that will be sequentially processed
opt_chunk_size(ctg) <- 0
plot(ctg, chunk = TRUE)
opt_chunk_size(ctg) <- 900
plot(ctg, chunk = TRUE)
Partial processing
It possible to flag some file that will not be processed but that will be used to load a buffer if required. In the following example only
the central files will be processed but the others one were not removed and they will be used to buffer the processed files.
ctg$processed <- FALSE
ctg$processed[c(19:20, 41:44, 49:50)] <- TRUE
plot(ctg)
Some practical examples
Example 1 - Raster
Load a collection Process each file sequentially. Returns a raster into R.
ctg <- readLAScatalog("path/to/las/files/")
hmean <- pixel_metrics(ctg, ~mean(Z), 20)
Example 2 - Raster
Load a collection Process each file sequentially. For each file write a raster on disk named after the name of the processed files. Returns a lightweight virtual raster mosaic.
ctg <- catalog("path/to/las/files/")
opt_output_files(ctg) <- "folder/where/to/store/outputs/dtm_{ORIGINALFILENAME}"
dtm <- rasterize_terrain(ctg, tin())
Example 3 - Raster
Load a single big file too big to be actually loaded in memory. Process small chunks of 100 x 100 meters at a time. Returns a raster into R.
ctg <- readLAScatalog("path/to/las/files/bigfile.las")
opt_chunk_size(ctg) <- 100
chm <- rasterize_canopy(ctg, p2r())
Example 4 - Tree detection
Load a collection. Process small chunks of 200 x 200 meters at a time. Each chunk is loaded with an extra 20 m buffer. Returns spatial points into R.
ctg <- readLAScatalog("path/to/las/files/")
opt_chunk_size(ctg) <- 200
opt_chunk_buffer(ctg) <- 20
ttops <- locate_trees(ctg, lmf(5))
Example 5 - Decimate
This is forbidden. The output would be too big.
ctg <- readLAScatalog("path/to/las/files/")
decimated <- decimate_points(ctg, homogenize(4))
Example 6 - Decimate
Load a collection. Process small chunks of 500 x 500 meter sequentially. For each chunk write a laz file on disk named after the coordinates of the chunk. Returns a lightweight LAScatalog. Note that the original collection has been retiled.
ctg <- readLAScatalog("path/to/las/files/")
opt_chunk_size(ctg) <- 500
opt_output_files(ctg) <- "folder/where/to/store/outputs/project_{XLEFT}_{YBOTTOM}_decimated"
opt_laz_compression(ctg) <- TRUE
decimated <- decimate_points(ctg, homogenize(4))
Example 7 - Clip
Load a collection. Load a shapefile of plot centers. Extract the plots. Returns a list of extracted point clouds in R.
ctg <- readLAScatalog("path/to/las/files/")
shp <- sf::st_read("plot_center.shp")
plots <- clip_roi(ctg, shp, radius = 11.2)
Example 8 - Clip
Load a collection. Load a shapefile of plot centers. Extract the plots and immediately write them into a file named after the coordinates of the plot and an attributes of the shapefile (here PLOTID if such an attribute exists in the shapefile). Returns a lightweight LAScatalog.
ctg <- readLAScatalog("path/to/las/files/")
shp <- sf::st_read("plot_center.shp")
opt_output_files(ctg) <- "folder/where/to/store/outputs/plot_{XCENTER}_{YCENTER}_{PLOTID}"
plots <- clip_roi(ctg, plot_centers, radius = 11.2)
Try the lidR package in your browser
Any scripts or data that you put into this service are public.
lidR documentation built on Sept. 11, 2024, 5:21 p.m.
knitr::opts_chunk$set( collapse = TRUE, comment = "#>", dev.args = list(pointsize = 10) ) options(rmarkdown.html_vignette.check_title = FALSE) library(lidR)
data = data.frame( Max.X = c(885228.88, 886993.96, 885260.93, 887025.96, 885292.94, 887056.88, 892199.94, 893265.54, 892229.99, 893295.15, 888759.96, 890524.95, 892259.98, 894025.98, 892289.96, 894055.93, 888790.91, 890554.98, 888820.95, 890585.99, 892319.96, 894084.97, 892349.89, 894114.29, 895250.23, 895094.78, 895044.96, 895053.55, 885323.96, 887087.95, 885355.95, 887119.96, 883657.85, 885387.95, 887150.97, 885419.98, 887182.95, 883688.44, 885442.91, 887193.9, 888851.96, 890615.97, 888882.94, 890646.97, 892379.94, 894127.84, 892409.97, 892676.58, 888913.92, 890676.93, 888944.86, 890707.98, 892439.95, 894124.59, 892469.86, 894232.94, 894786.68, 888958.83, 890713.51, 892476.43, 894239.97, 894786.07), Min.X = c(885022.37, 885204.73, 885027.52, 885229.03, 885040.86, 885261.03, 891503.09, 892198.69, 891501.42, 892200.07, 886970.07, 888735.55, 891499.96, 892230.05, 890521.99, 892260.01, 886994.05, 888760.09, 887026.07, 888791.01, 890525.05, 892290.04, 890555.01, 892320.12, 894002.98, 894026.02, 894056.02, 894085.03, 885051.45, 885293.03, 885063.29, 885324.03, 883166.09, 885072.16, 885356.09, 883642.36, 885388.15, 883180.23, 883658.11, 885420.02, 887057.07, 888821.02, 887088.11, 888852.03, 890586.03, 892350.02, 890616.07, 892380.01, 887120.07, 888883.03, 887151.11, 888914.02, 890647.06, 892410.06, 890677.07, 892440.07, 894209.19, 887183.07, 888945.12, 890708.03, 892470.16, 894233.07), Max.Y = c(630219.48, 630214.96, 631609.95, 631604.97, 633001.65, 632995.99, 625898.35, 625882.94, 627289.82, 627273.89, 630174.88, 630134.94, 628681.66, 628664.99, 630094.95, 630057.95, 631564.98, 631524.94, 632955.82, 632915.99, 631486.9, 631447.96, 632876.93, 632838.96, 628627.89, 630019.93, 631410.97, 631740.88, 634393.05, 634386.96, 635786.24, 635779.96, 638613.36, 637176.84, 637169.92, 638601.99, 638560.96, 639938.36, 639926.95, 639558.31, 634346.93, 634307.92, 635739.92, 635699.92, 634268.97, 634229.95, 635659.89, 635622.88, 637129.84, 637089.93, 638520.94, 638481.91, 637051.99, 637012.92, 638442.98, 638403.94, 638366.87, 639177.04, 639133.74, 638702.56, 638702.56, 638702.56), Min.Y = c(629157.18, 629099.31, 630215.04, 630175.05, 631605.02, 631565.05, 625816.52, 625793.6, 625883.01, 625860.81, 629036.82, 629017.72, 627274.01, 627251.36, 628665.04, 628628.01, 630135.08, 630095.02, 631525.01, 631487.19, 630058.02, 630020.05, 631448.08, 631411.03, 627506.32, 628612.41, 629999.84, 631390.38, 632996.06, 632956.04, 634387.01, 634347.01, 637939.24, 635780.07, 635740.05, 637170.11, 637130.14, 638602.13, 638561.04, 638521.07, 632916.05, 632877.04, 634308.06, 634269.04, 632839.06, 632801.04, 634230.04, 634223.9, 635700.07, 635660.11, 637090.03, 637052.15, 635623.06, 635619.13, 637013.1, 636979.71, 637259.33, 638482.01, 638443.02, 638404.08, 638367.11, 638355.37), Min.Z = c(325.12, 251.48, 244.68, 286.7, 338.86, 320.68, 118.08, 60.69, -5.01, -7.58, 225.29, 252, 87.3, 41.7, 115.01, 28.77, 205.11, 200.85, 200.54, 169.5, 90.64, 19.72, 126.04, 28.6, 41.98, 43.15, 7.74, 6.7, 199.26, 190.02, 284.92, 216.16, 218.14, 318.93, 220.21, 218.04, 137.31, 218.13, 217.34, 190.42, 207.62, 113.84, 118.18, 141.52, 92, 52.5, 91.2, 77.92, 156.57, 89.53, 108.83, 93.98, 23.45, 1.64, 33.22, 3.29, 0.61, 108.03, 208.38, 121.18, 58.83, 0.95), Max.Z = c(418.46, 990.54, 409.06, 1021.87, 996.42, 1005.02, 173.77, 393.97, 836.52, 820.98, 414.2, 936.47, 792.95, 822.51, 777.31, 837.87, 419.15, 741.84, 907.2, 872.27, 898.53, 822.53, 846.77, 740.65, 826.61, 890.21, 828.86, 680.32, 390.31, 997.2, 965.55, 969.24, 249.34, 849.5, 950.2, 848.64, 904.1, 880, 827.92, 888.34, 462.88, 906.61, 440.83, 887.34, 860.37, 747.1, 808.75, 194.76, 734.21, 838.34, 834.76, 758.91, 771.76, 670.1, 810.94, 761.53, 109.26, 303.94, 349.94, 799.8, 737.01, 593.91), Version.Major = 1, Version.Minor = 2, Point.Data.Format.ID = 1, filename = paste0("path/to/las/files/file", 1:62, ".las") ) geom <- lapply(1:nrow(data), function(i) { mtx <- matrix(c(data$Min.X[i], data$Max.X[i], data$Min.Y[i], data$Max.Y[i])[c(1, 1, 2, 2, 1, 3, 4, 4, 3, 3)], ncol = 2) sf::st_polygon(list(mtx)) }) geom <-sf::st_sfc(geom) sf::st_crs(geom) <- 26917 data <- sf::st_set_geometry(data, geom) ctg <- new("LAScatalog") ctg@data <- data
A LAScatalog class is a representation in R of a las file or a collection of las files not loaded in memory. Indeed, a regular computer cannot load the entire point cloud in R if it covers a broad area. For very high density datasets it can even fail loading a single file (see also the "LAS formal class" vignette). In lidR, we use a LAScatalog to process datasets that cannot fit in memory.
Build a LAScatalog object reading a folder of las files
ctg <- readLAScatalog("path/to/las/files/") # or ctg <- readLAScatalog("path/to/las/files/big_file.las")
ctg
Basic structure of a LAScatalog object
A LAScatalog contains a sf object with POLYGON geometries plus some extra slots that store information relative to how the LAScatalog will be processed.
The slot data of a LAScatalog object contains the sf object with the most important information read from the header of .las or .laz files. Reading only the header of the file provides an overview of the content of the files very quickly without actually loading the point cloud. The columns of the table are named after the LAS specification version 1.4
The other slots are well documented in the documentation help("LAScatalog-class") so they are not described in this vignette.
Allowed and non-allowed manipulation of a LAScatalog object
A LAScatalog purpose is not to manipulate spatial data in R. The purpose of a LAScatalog is to represent a set of existing las/laz files. Thus a LAScatalog cannot be modified because it must be related to the actual content of the files. The following throws an error:
ctg$Min.Z <- 0
Obviously it is always possible to modify an R object by bypassing such simple restrictions. In this case the user will break something internally and a correct output is not guaranteed.
However it is possible to add and modify the attributes using a name that is not reserved. The following is allowed:
ctg$newattribute <- 0
Validation of LAScatalog object
Users commonly report bugs arising from the fact that the point cloud is invalid. This is why we introduced the function las_check() to perform an inspection of the LAScatalog objects. This function checks if a LAScatalog object is consistent (files are all of the same type for example). For example, it may happen that a collection mixes files of type 1 with files of type 3 or files with different scale factors.
las_check(ctg)
The function las_check() when applied to a LAScatalog does not perform a deep inspection of the point cloud unlike when applied to a LAS object. Indeed the point cloud is not actually read.
Display a LAScatalog object
lidR provides a simple plot() function to plot a LAScatalog object:
plot(ctg)
The option mapview = TRUE displays the LAScatalog on an interactive map with pan and zoom and allows the addition of a satellite map in the background. It uses the package mapview internally. It is often useful to check if the CRS of the file are properly registered. The epsg codes recorded in the las files appear to be sometime incorrect, according to our own experience.
plot(ctg, mapview = TRUE, map.type = "Esri.WorldImagery")

Using a sf object to store the attributes of the las file it is easy to display metadata of the files. In the following we can immediately see that the catalog is not normalized and is likely to contain outliers:
plot(ctg["Min.Z"])
Apply lidR functions on a LAScatalog
Most of lidR functions are compatible with a LAScatalog and work almost like with a single point cloud loaded in memory. In the following example we use the function pixel_metrics() to compute the mean elevation of the points. The output is a continuous wall-to-wall raster. It works exactly as if the input was a LAS object.
hmean <- pixel_metrics(ctg, mean(Z), 20)
However, processing a LAScatalog usually requires some tuning of the processing options to get better control of the computation. Indeed, if the catalog is huge the output is likely to be huge as well, and maybe the output cannot fit in the R memory. For example, normalize_height() throws an error if used 'as is' without tuning the processing options. Using normalize_height() like in the following example the expected output would be a huge point cloud loaded in memory. The lidR package forbids such a call:
output <- normalize_height(ctg, tin())
Instead, one can use the processing option opt_output_files(). Processing options drive how the big files are split in small chunks and how the outputs are either returned into R or written on disk into files.
opt_output_files(ctg) <- "folder/where/to/store/outputs/{ORIGINALFILENAME}_normalized" output <- normalize_height(ctg, tin())
Here the output is not a point cloud but a LAScatalog pointing to the newly created files. The user can check how the collection will be processed by calling summary
summary(ctg)
Also the plot function can displays the chunks pattern i.e. how the dataset is split into small chunks that will be sequentially processed
opt_chunk_size(ctg) <- 0 plot(ctg, chunk = TRUE) opt_chunk_size(ctg) <- 900 plot(ctg, chunk = TRUE)
Partial processing
It possible to flag some file that will not be processed but that will be used to load a buffer if required. In the following example only the central files will be processed but the others one were not removed and they will be used to buffer the processed files.
ctg$processed <- FALSE ctg$processed[c(19:20, 41:44, 49:50)] <- TRUE plot(ctg)
Some practical examples
Example 1 - Raster
Load a collection Process each file sequentially. Returns a raster into R.
ctg <- readLAScatalog("path/to/las/files/") hmean <- pixel_metrics(ctg, ~mean(Z), 20)
Example 2 - Raster
Load a collection Process each file sequentially. For each file write a raster on disk named after the name of the processed files. Returns a lightweight virtual raster mosaic.
ctg <- catalog("path/to/las/files/") opt_output_files(ctg) <- "folder/where/to/store/outputs/dtm_{ORIGINALFILENAME}" dtm <- rasterize_terrain(ctg, tin())
Example 3 - Raster
Load a single big file too big to be actually loaded in memory. Process small chunks of 100 x 100 meters at a time. Returns a raster into R.
ctg <- readLAScatalog("path/to/las/files/bigfile.las") opt_chunk_size(ctg) <- 100 chm <- rasterize_canopy(ctg, p2r())
Example 4 - Tree detection
Load a collection. Process small chunks of 200 x 200 meters at a time. Each chunk is loaded with an extra 20 m buffer. Returns spatial points into R.
ctg <- readLAScatalog("path/to/las/files/") opt_chunk_size(ctg) <- 200 opt_chunk_buffer(ctg) <- 20 ttops <- locate_trees(ctg, lmf(5))
Example 5 - Decimate
This is forbidden. The output would be too big.
ctg <- readLAScatalog("path/to/las/files/") decimated <- decimate_points(ctg, homogenize(4))
Example 6 - Decimate
Load a collection. Process small chunks of 500 x 500 meter sequentially. For each chunk write a laz file on disk named after the coordinates of the chunk. Returns a lightweight LAScatalog. Note that the original collection has been retiled.
ctg <- readLAScatalog("path/to/las/files/") opt_chunk_size(ctg) <- 500 opt_output_files(ctg) <- "folder/where/to/store/outputs/project_{XLEFT}_{YBOTTOM}_decimated" opt_laz_compression(ctg) <- TRUE decimated <- decimate_points(ctg, homogenize(4))
Example 7 - Clip
Load a collection. Load a shapefile of plot centers. Extract the plots. Returns a list of extracted point clouds in R.
ctg <- readLAScatalog("path/to/las/files/") shp <- sf::st_read("plot_center.shp") plots <- clip_roi(ctg, shp, radius = 11.2)
Example 8 - Clip
Load a collection. Load a shapefile of plot centers. Extract the plots and immediately write them into a file named after the coordinates of the plot and an attributes of the shapefile (here PLOTID if such an attribute exists in the shapefile). Returns a lightweight LAScatalog.
ctg <- readLAScatalog("path/to/las/files/") shp <- sf::st_read("plot_center.shp") opt_output_files(ctg) <- "folder/where/to/store/outputs/plot_{XCENTER}_{YCENTER}_{PLOTID}" plots <- clip_roi(ctg, plot_centers, radius = 11.2)
Try the lidR package in your browser
Any scripts or data that you put into this service are public.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
