Correlation Plots
In metasnf: Meta Clustering with Similarity Network Fusion
knitr::opts_chunk$set(echo = TRUE)
options(crayon.enabled = FALSE, cli.num_colors = 0)
Download a copy of the vignette to follow along here: correlation_plots.Rmd
In this vignette, we go through how you can visualize associations between the features included in your analyses.
Data set-up
library(metasnf)
# We'll just use the first few columns for this demo
cort_sa_minimal <- cort_sa[, 1:5]
# And one more mock categorical feature for demonstration purposes
city <- fav_colour
city$"city" <- sample(
c("toronto", "montreal", "vancouver"),
size = nrow(city),
replace = TRUE
)
city <- city |> dplyr::select(-"colour")
# Make sure to throw in all the data you're interested in visualizing for this
# data_list, including out-of-model measures and confounding features.
dl <- data_list(
list(cort_sa_minimal, "cortical_sa", "neuroimaging", "continuous"),
list(income, "household_income", "demographics", "ordinal"),
list(pubertal, "pubertal_status", "demographics", "continuous"),
list(fav_colour, "favourite_colour", "demographics", "categorical"),
list(city, "city", "demographics", "categorical"),
list(anxiety, "anxiety", "behaviour", "ordinal"),
list(depress, "depressed", "behaviour", "ordinal"),
uid = "unique_id"
)
summary(dl)
# This matrix contains all the pairwise association p-values
assoc_pval_matrix <- calc_assoc_pval_matrix(dl)
assoc_pval_matrix[1:3, 1:3]
Heatmaps
Here's what a basic heatmap looks like:
ap_heatmap <- assoc_pval_heatmap(assoc_pval_matrix)
save_heatmap(
ap_heatmap,
"assoc_pval_heatmap.png",
width = 650,
height = 500,
res = 100
)
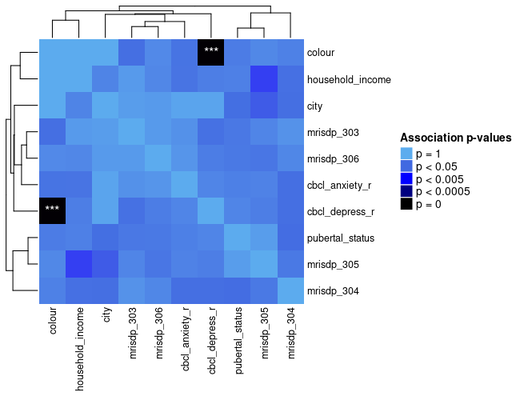
Most of this data was generated randomly, but the "colour" feature is really just a categorical mapping of "cbcl_depress_r".
You can draw attention to confounding features and/or any out of model measures by specifying their names as shown below.
ap_heatmap2 <- assoc_pval_heatmap(
assoc_pval_matrix,
confounders = list(
"Colour" = "colour",
"Pubertal Status" = "pubertal_status"
),
out_of_models = list(
"City" = "city"
)
)
save_heatmap(
ap_heatmap2,
"assoc_pval_heatmap2.png",
width = 680,
height = 500,
res = 100
)
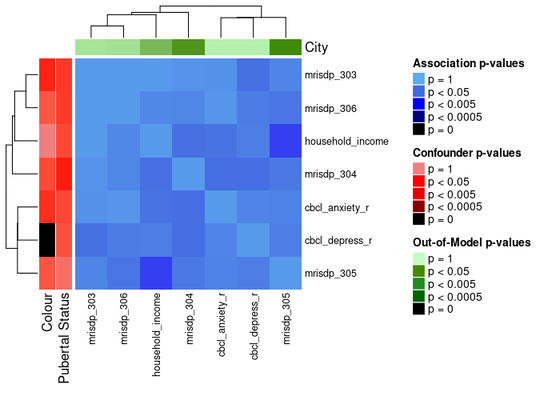
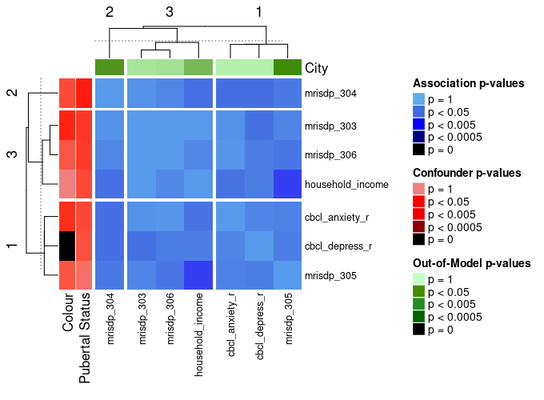
The ComplexHeatmap package offers functionality for splitting heatmaps into slices.
One way to do the slices is by clustering the heatmap with k-means:
ap_heatmap3 <- assoc_pval_heatmap(
assoc_pval_matrix,
confounders = list(
"Colour" = "colour",
"Pubertal Status" = "pubertal_status"
),
out_of_models = list(
"City" = "city"
),
row_km = 3,
column_km = 3
)
save_heatmap(
ap_heatmap3,
"assoc_pval_heatmap3.png",
width = 680,
height = 500,
res = 100
)
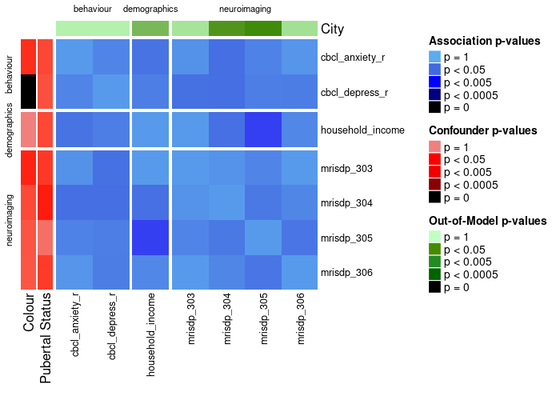
Another way to divide the heatmap is by feature domain.
This can be done by providing a data_list with all the features in the assoc_pval_matrix and setting split_by_domain to TRUE.
ap_heatmap4 <- assoc_pval_heatmap(
assoc_pval_matrix,
confounders = list(
"Colour" = "colour",
"Pubertal Status" = "pubertal_status"
),
out_of_models = list(
"City" = "city"
),
dl = data_list,
split_by_domain = TRUE
)
save_heatmap(
ap_heatmap4,
"assoc_pval_heatmap4.png",
width = 700,
height = 500,
res = 100
)
Try the metasnf package in your browser
Any scripts or data that you put into this service are public.
metasnf documentation built on June 8, 2025, 12:47 p.m.
knitr::opts_chunk$set(echo = TRUE)
options(crayon.enabled = FALSE, cli.num_colors = 0)
Download a copy of the vignette to follow along here: correlation_plots.Rmd
In this vignette, we go through how you can visualize associations between the features included in your analyses.
Data set-up
library(metasnf) # We'll just use the first few columns for this demo cort_sa_minimal <- cort_sa[, 1:5] # And one more mock categorical feature for demonstration purposes city <- fav_colour city$"city" <- sample( c("toronto", "montreal", "vancouver"), size = nrow(city), replace = TRUE ) city <- city |> dplyr::select(-"colour") # Make sure to throw in all the data you're interested in visualizing for this # data_list, including out-of-model measures and confounding features. dl <- data_list( list(cort_sa_minimal, "cortical_sa", "neuroimaging", "continuous"), list(income, "household_income", "demographics", "ordinal"), list(pubertal, "pubertal_status", "demographics", "continuous"), list(fav_colour, "favourite_colour", "demographics", "categorical"), list(city, "city", "demographics", "categorical"), list(anxiety, "anxiety", "behaviour", "ordinal"), list(depress, "depressed", "behaviour", "ordinal"), uid = "unique_id" ) summary(dl) # This matrix contains all the pairwise association p-values assoc_pval_matrix <- calc_assoc_pval_matrix(dl) assoc_pval_matrix[1:3, 1:3]
Heatmaps
Here's what a basic heatmap looks like:
ap_heatmap <- assoc_pval_heatmap(assoc_pval_matrix)
save_heatmap( ap_heatmap, "assoc_pval_heatmap.png", width = 650, height = 500, res = 100 )

Most of this data was generated randomly, but the "colour" feature is really just a categorical mapping of "cbcl_depress_r".
You can draw attention to confounding features and/or any out of model measures by specifying their names as shown below.
ap_heatmap2 <- assoc_pval_heatmap( assoc_pval_matrix, confounders = list( "Colour" = "colour", "Pubertal Status" = "pubertal_status" ), out_of_models = list( "City" = "city" ) )
save_heatmap( ap_heatmap2, "assoc_pval_heatmap2.png", width = 680, height = 500, res = 100 )

The ComplexHeatmap package offers functionality for splitting heatmaps into slices. One way to do the slices is by clustering the heatmap with k-means:
ap_heatmap3 <- assoc_pval_heatmap( assoc_pval_matrix, confounders = list( "Colour" = "colour", "Pubertal Status" = "pubertal_status" ), out_of_models = list( "City" = "city" ), row_km = 3, column_km = 3 )
save_heatmap( ap_heatmap3, "assoc_pval_heatmap3.png", width = 680, height = 500, res = 100 )

Another way to divide the heatmap is by feature domain.
This can be done by providing a data_list with all the features in the assoc_pval_matrix and setting split_by_domain to TRUE.
ap_heatmap4 <- assoc_pval_heatmap( assoc_pval_matrix, confounders = list( "Colour" = "colour", "Pubertal Status" = "pubertal_status" ), out_of_models = list( "City" = "city" ), dl = data_list, split_by_domain = TRUE )
save_heatmap( ap_heatmap4, "assoc_pval_heatmap4.png", width = 700, height = 500, res = 100 )

Try the metasnf package in your browser
Any scripts or data that you put into this service are public.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
