Similarity Matrices
In metasnf: Meta Clustering with Similarity Network Fusion
knitr::opts_chunk$set(echo = TRUE)
options(crayon.enabled = FALSE, cli.num_colors = 0)
Download a copy of the vignette to follow along here: similarity_matrix_heatmaps.Rmd
This vignette walks through usage of similarity_matrix_heatmap to visualize the final similarity matrix produced by a run of SNF and how that matrix associates with other patient attributes.
Data set-up
library(metasnf)
# Generate data_list
my_dl <- data_list(
list(
data = expression_df,
name = "expression_data",
domain = "gene_expression",
type = "continuous"
),
list(
data = methylation_df,
name = "methylation_data",
domain = "gene_methylation",
type = "continuous"
),
list(
data = gender_df,
name = "gender",
domain = "demographics",
type = "categorical"
),
list(
data = diagnosis_df,
name = "diagnosis",
domain = "clinical",
type = "categorical"
),
list(
data = age_df,
name = "age",
domain = "demographics",
type = "discrete"
),
uid = "patient_id"
)
set.seed(42)
my_sc <- snf_config(
my_dl,
n_solutions = 1,
max_k = 40
)
sol_df <- batch_snf(
my_dl,
my_sc,
return_sim_mats = TRUE
)
similarity_matrices <- sim_mats_list(sol_df)
# The first (and only) similarity matrix:
similarity_matrix <- similarity_matrices[[1]]
# The first (and only) cluster solution:
cluster_solution <- t(sol_df)
Visualize similarity matrices sorted by cluster label
similarity_matrix_heatmap is a wrapper for ComplexHeatmap::Heatmap, but with some convenient default transformations and parameters for viewing a similarity matrix.
similarity_matrix_hm <- similarity_matrix_heatmap(
similarity_matrix = similarity_matrix,
cluster_solution = cluster_solution,
heatmap_height = grid::unit(10, "cm"),
heatmap_width = grid::unit(10, "cm")
)
save_heatmap(
heatmap = similarity_matrix_hm,
path = "vignettes/similarity_matrix_heatmap.png",
width = 410,
height = 330,
res = 80
)
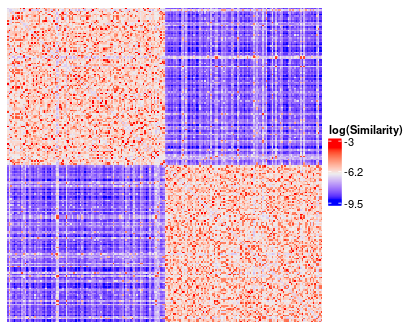
The default transformations include plotting log(Similarity) rather than the default similarity matrix as well as rescaling the diagonal of the matrix to the average value of the off-diagonals.
Additionally, the similarity matrix gets reordered according to the provided cluster solution.
Annotations
One piece of functionality provided by ComplexHeatmap::Heatmap() is the ability to supply visual annotations along the rows and columns of a heatmap.
You can always build annotations using the standard approaches outline in the ComplexHeatmap Complete Reference.
In addition to that, this package offers some convenient functionality to specify regular heatmap annotations and barplot annotations directly through a provided data frame or data_list (or both).
In the example below, we make use of data supplied through a data list.
annotated_sm_hm <- similarity_matrix_heatmap(
similarity_matrix = similarity_matrix,
cluster_solution = cluster_solution,
scale_diag = "mean",
log_graph = TRUE,
data = my_dl,
left_hm = list(
"Diagnosis" = "diagnosis"
),
top_hm = list(
"Gender" = "gender"
),
top_bar = list(
"Age" = "age"
),
annotation_colours = list(
Diagnosis = c(
"definite asthma" = "red3",
"possible asthma" = "pink1",
"no asthma" = "bisque1"
),
Gender = c(
"female" = "purple",
"male" = "lightgreen"
)
),
heatmap_height = grid::unit(10, "cm"),
heatmap_width = grid::unit(10, "cm")
)
save_heatmap(
heatmap = annotated_sm_hm,
path = "vignettes/annotated_sm_heatmap.png",
width = 500,
height = 440,
res = 80
)
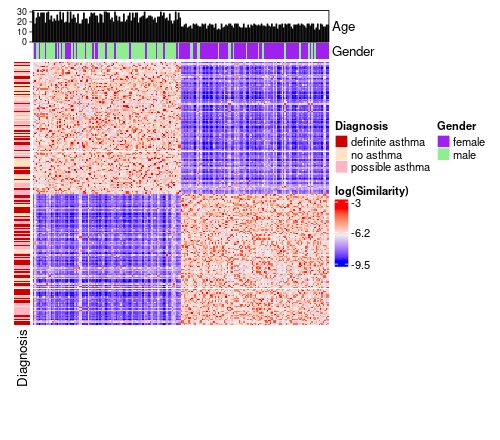
The colours red3, pink1, etc. are built-in R colours that you can browse by calling colours().
For reference, the code below shows how you would achieve these annotations using standard ComplexHeatmap syntax.
merged_df <- as.data.frame(my_dl)
order <- sort(cluster_solution[, 2], index.return = TRUE)$"ix"
merged_df <- merged_df[order, ]
top_annotations <- ComplexHeatmap::HeatmapAnnotation(
Age = ComplexHeatmap::anno_barplot(merged_df$"age"),
Gender = merged_df$"gender",
col = list(
Gender = c(
"female" = "purple",
"male" = "lightgreen"
)
),
show_legend = TRUE
)
left_annotations <- ComplexHeatmap::rowAnnotation(
Diagnosis = merged_df$"diagnosis",
col = list(
Diagnosis = c(
"definite asthma" = "red3",
"possible asthma" = "pink1",
"no asthma" = "bisque1"
)
),
show_legend = TRUE
)
similarity_matrix_heatmap(
similarity_matrix = similarity_matrix,
cluster_solution = cluster_solution,
scale_diag = "mean",
log_graph = TRUE,
data = merged_df,
top_annotation = top_annotations,
left_annotation = left_annotations
)
Take a look at the ComplexHeatmap Complete Reference to learn more about what is possible with this package.
More on sorting
Be aware that the ordering of both your data and your similarity matrix will be influenced if you supply values for the cluster_solution or order parameters.
If you don't think your data is lining up properly, consider manually making sure your similarity_matrix rows and columns are sorted to your preference (e.g., based on cluster) and that the order of your data matches.
This will be easier to do with a data frame than with a data_list, as the data_list forces patients to be sorted by their unique IDs upon generation.
Try the metasnf package in your browser
Any scripts or data that you put into this service are public.
metasnf documentation built on June 8, 2025, 12:47 p.m.
knitr::opts_chunk$set(echo = TRUE)
options(crayon.enabled = FALSE, cli.num_colors = 0)
Download a copy of the vignette to follow along here: similarity_matrix_heatmaps.Rmd
This vignette walks through usage of similarity_matrix_heatmap to visualize the final similarity matrix produced by a run of SNF and how that matrix associates with other patient attributes.
Data set-up
library(metasnf) # Generate data_list my_dl <- data_list( list( data = expression_df, name = "expression_data", domain = "gene_expression", type = "continuous" ), list( data = methylation_df, name = "methylation_data", domain = "gene_methylation", type = "continuous" ), list( data = gender_df, name = "gender", domain = "demographics", type = "categorical" ), list( data = diagnosis_df, name = "diagnosis", domain = "clinical", type = "categorical" ), list( data = age_df, name = "age", domain = "demographics", type = "discrete" ), uid = "patient_id" ) set.seed(42) my_sc <- snf_config( my_dl, n_solutions = 1, max_k = 40 ) sol_df <- batch_snf( my_dl, my_sc, return_sim_mats = TRUE ) similarity_matrices <- sim_mats_list(sol_df) # The first (and only) similarity matrix: similarity_matrix <- similarity_matrices[[1]] # The first (and only) cluster solution: cluster_solution <- t(sol_df)
Visualize similarity matrices sorted by cluster label
similarity_matrix_heatmap is a wrapper for ComplexHeatmap::Heatmap, but with some convenient default transformations and parameters for viewing a similarity matrix.
similarity_matrix_hm <- similarity_matrix_heatmap( similarity_matrix = similarity_matrix, cluster_solution = cluster_solution, heatmap_height = grid::unit(10, "cm"), heatmap_width = grid::unit(10, "cm") )
save_heatmap( heatmap = similarity_matrix_hm, path = "vignettes/similarity_matrix_heatmap.png", width = 410, height = 330, res = 80 )

The default transformations include plotting log(Similarity) rather than the default similarity matrix as well as rescaling the diagonal of the matrix to the average value of the off-diagonals. Additionally, the similarity matrix gets reordered according to the provided cluster solution.
Annotations
One piece of functionality provided by ComplexHeatmap::Heatmap() is the ability to supply visual annotations along the rows and columns of a heatmap.
You can always build annotations using the standard approaches outline in the ComplexHeatmap Complete Reference. In addition to that, this package offers some convenient functionality to specify regular heatmap annotations and barplot annotations directly through a provided data frame or data_list (or both).
In the example below, we make use of data supplied through a data list.
annotated_sm_hm <- similarity_matrix_heatmap( similarity_matrix = similarity_matrix, cluster_solution = cluster_solution, scale_diag = "mean", log_graph = TRUE, data = my_dl, left_hm = list( "Diagnosis" = "diagnosis" ), top_hm = list( "Gender" = "gender" ), top_bar = list( "Age" = "age" ), annotation_colours = list( Diagnosis = c( "definite asthma" = "red3", "possible asthma" = "pink1", "no asthma" = "bisque1" ), Gender = c( "female" = "purple", "male" = "lightgreen" ) ), heatmap_height = grid::unit(10, "cm"), heatmap_width = grid::unit(10, "cm") )
save_heatmap( heatmap = annotated_sm_hm, path = "vignettes/annotated_sm_heatmap.png", width = 500, height = 440, res = 80 )

The colours red3, pink1, etc. are built-in R colours that you can browse by calling colours().
For reference, the code below shows how you would achieve these annotations using standard ComplexHeatmap syntax.
merged_df <- as.data.frame(my_dl) order <- sort(cluster_solution[, 2], index.return = TRUE)$"ix" merged_df <- merged_df[order, ] top_annotations <- ComplexHeatmap::HeatmapAnnotation( Age = ComplexHeatmap::anno_barplot(merged_df$"age"), Gender = merged_df$"gender", col = list( Gender = c( "female" = "purple", "male" = "lightgreen" ) ), show_legend = TRUE ) left_annotations <- ComplexHeatmap::rowAnnotation( Diagnosis = merged_df$"diagnosis", col = list( Diagnosis = c( "definite asthma" = "red3", "possible asthma" = "pink1", "no asthma" = "bisque1" ) ), show_legend = TRUE ) similarity_matrix_heatmap( similarity_matrix = similarity_matrix, cluster_solution = cluster_solution, scale_diag = "mean", log_graph = TRUE, data = merged_df, top_annotation = top_annotations, left_annotation = left_annotations )
Take a look at the ComplexHeatmap Complete Reference to learn more about what is possible with this package.
More on sorting
Be aware that the ordering of both your data and your similarity matrix will be influenced if you supply values for the cluster_solution or order parameters.
If you don't think your data is lining up properly, consider manually making sure your similarity_matrix rows and columns are sorted to your preference (e.g., based on cluster) and that the order of your data matches.
This will be easier to do with a data frame than with a data_list, as the data_list forces patients to be sorted by their unique IDs upon generation.
Try the metasnf package in your browser
Any scripts or data that you put into this service are public.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
