inst/examples/tutorial.md
In rfigshare: An R Interface to 'figshare'
title: rfigshare - Share and explore figures, data, and publications on FigShare using R

Obtaining your API keys
There is a nice video introduction to creating applications for the API on the figshare blog. The following tutorial provides a simple walkthrough of how to go about getting your figshare API keys set up so that you can use the rfigshare package.
Create a user account on FigShare and log in. From your homepage, select "Applications" from the drop-down menu,
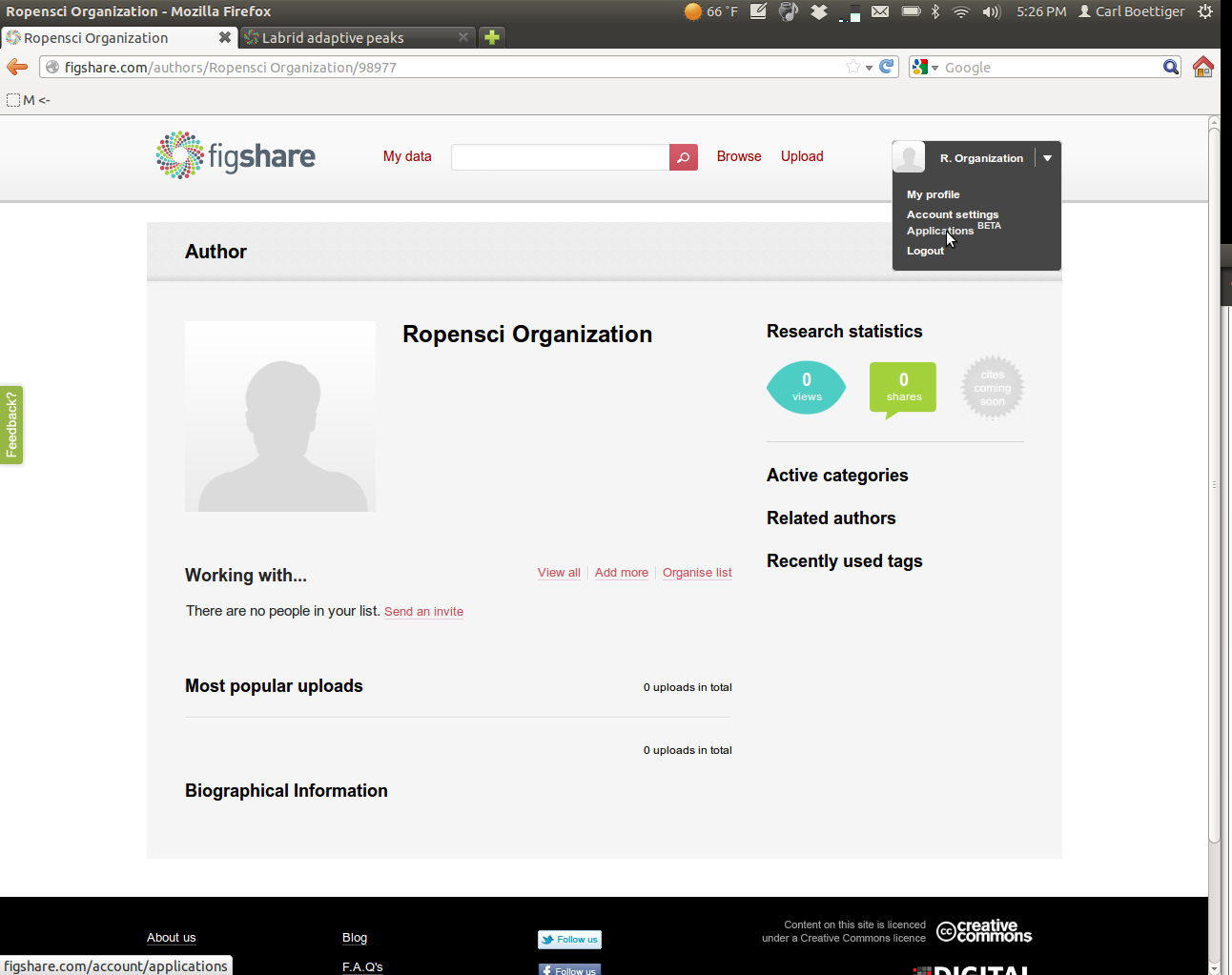
Create a new application:

Enter in the following information:
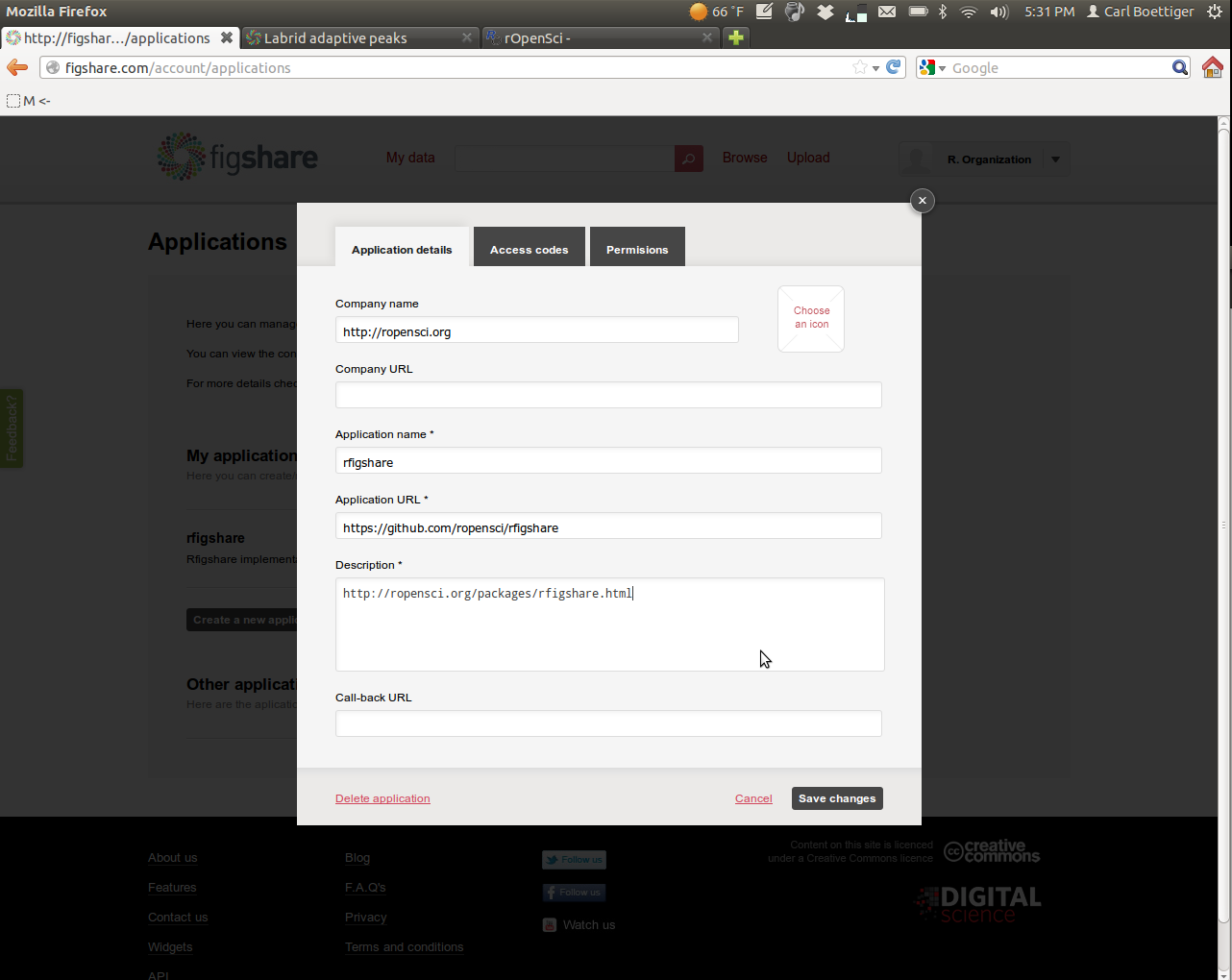
Then navigate over to the permissions tab. To get the most out of rfigshare you'll want to enable all permissions:
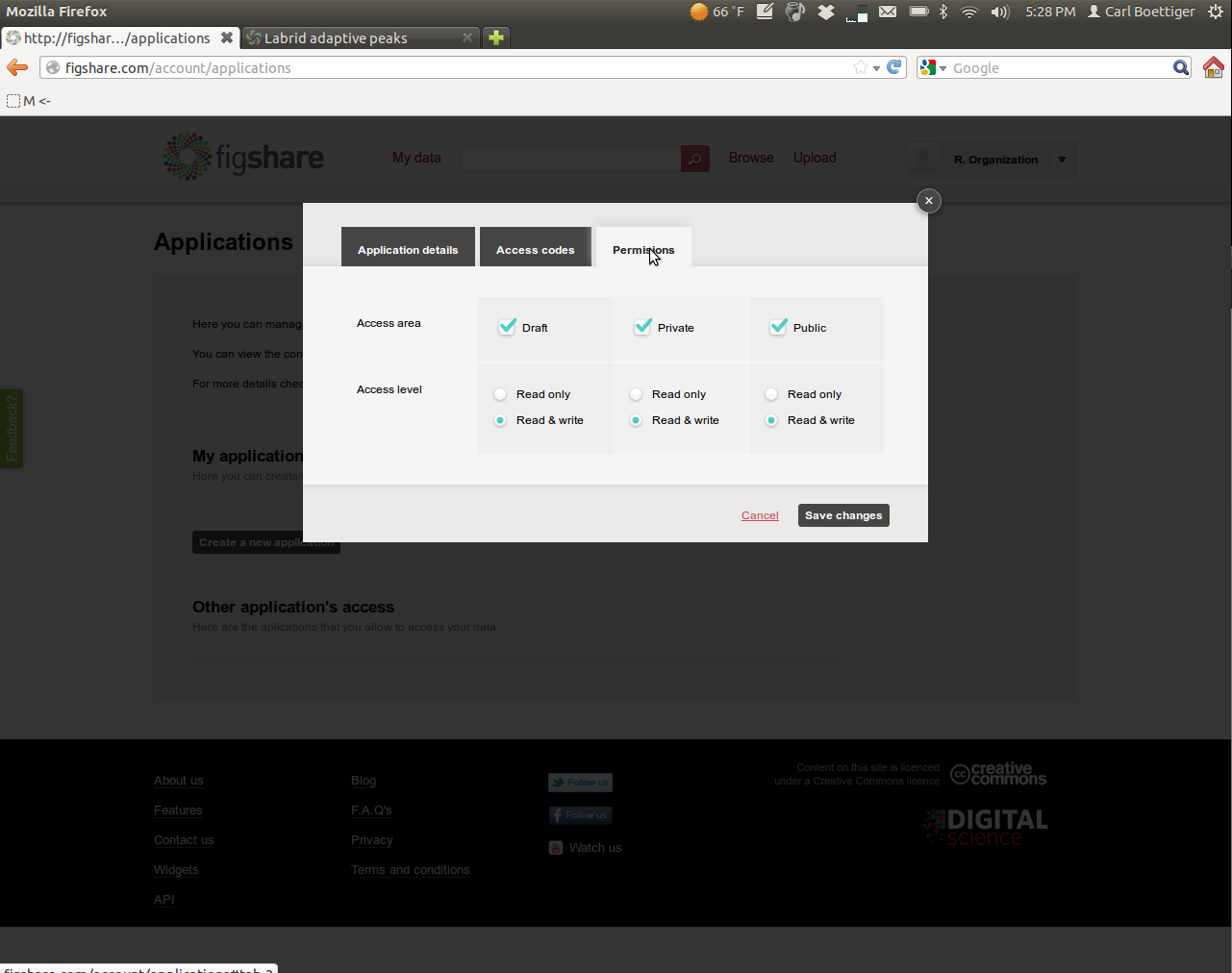
Save the new settings, and then open the application again (View/Edit menu) and click on the "Access Codes" tab.

Record each if the keys into R as follows. You might want to put this bit of R code into your .Rprofile to avoid entering it each time in the future:
options(FigshareKey = "qMDabXXXXXXXXXXXXXXXXX")
options(FigsharePrivateKey = "zQXXXXXXXXXXXXXXXXXXXX")
options(FigshareToken = "SrpxabQXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX")
options(FigsharePrivateToken = "yqXXXXXXXXXXXXXXXXXXXX")
Installing the R package
Now that we have the API credentials in place, actually using rfigshare is pretty easy. Install the latest version of package directly from Github using:
require(devtools)
install_github("rfigshare", "ropensci")
Using rfigshare
Try authenticating with your credentials:
require(rfigshare)
fs_auth()
Try a search for an author:
fs_author_search("Boettiger")
text_content() deprecated. Use parsed_content(x, as = 'parsed')
[[1]]
[[1]]$id
[1] "96387"
[[1]]$fname
[1] "Carl"
[[1]]$lname
[1] "Boettiger"
[[1]]$full_name
[1] "Carl Boettiger"
[[1]]$job_title
[1] ""
[[1]]$description
[1] ""
[[1]]$facebook
[1] ""
[[1]]$twitter
[1] ""
[[1]]$active
[1] 1
Try creating your own content:
id <- fs_create("Test title", "description of test", "dataset")
text_content() deprecated. Use parsed_content(x, as = 'parsed')
Your article has been created! Your id number is 105137
This creates an article with the essential metadata information. We can now upload the dataset to this newly created figshare object using fs_upload. For the purposes of this example, we'll just upload one of R's built-in datasets:
data(mtcars)
write.csv(mtcars, "mtcars.csv")
fs_upload(id, "mtcars.csv")
Not that we must pass the id number of our the newly created figshare object as the first argument. Similar functions let us add additional authors, tags, categories, and links, e.g.
fs_add_tags(id, "demo")
Minimal metadata includes title, description, type, and at least one tag and one category. We can add categories using either the category id or the name of the category, but it must be one of the preset categories available. We can ask the API for a list of all the categories:
fs_category_list()
id name
1 89 Aerospace Engineering
2 88 Agricultural Engineering
3 102 Algebra
4 10 Anatomy
5 25 Anthropology
6 77 Applied Computer Science
7 107 Applied Physics
8 127 Archaeology
9 94 Art
10 58 Astrophysics
11 78 Atmospheric Sciences
12 108 Atomic Physics
13 11 Behavioral neuroscience
14 4 Biochemistry
15 125 Bioinformatics
16 42 Biological engineering
17 1 Biophysics
18 21 Biotechnology
19 65 Botany
20 64 Cancer
21 12 Cell biology
22 109 Computational Physics
23 20 Computer Engineering
24 110 Condensed Matter Physics
25 57 Cosmology
26 66 Crystallography
27 95 Design
28 61 Developmental Biology
29 39 Ecology
30 27 Economics
31 129 Education
32 117 Entropy
33 31 Environmental chemistry
34 34 Environmental science
35 24 Evolutionary biology
36 56 Galactic Astronomy
37 118 General Relativity
38 90 Genetic Engineering
39 13 Genetics
40 32 Geochemistry
41 17 Geography
42 29 Geology
43 103 Geometry
44 79 Geophysics
45 128 Hematology
46 126 History
47 80 Hydrology
48 46 Immunology
49 69 Inorganic Chemistry
50 97 Law
51 35 Limnology
52 98 Literature
53 119 M-Theory
54 62 Marine Biology
55 91 Mechanical Engineering
56 111 Mechanics
57 7 Medicine
58 122 Mental Health
59 8 Microbiology
60 82 Mineralogy
61 14 Molecular biology
62 70 Molecular Physics
63 15 Neuroscience
64 71 Nuclear Chemistry
65 92 Nuclear Engineering
66 36 Oceanography
67 37 Organic chemistry
68 84 Paleoclimatology
69 28 Paleontology
70 85 Palynology
71 63 Parasitology
72 112 Particle Physics
73 99 Performing Arts
74 19 Pharmacology
75 100 Philosophy
76 86 Physical Geography
77 16 Physiology
78 54 Planetary Geology
79 59 Planetary Science
80 113 Plasma Physics
81 104 Probability
82 114 Quantum Mechanics
83 73 Radiochemistry
84 106 Science Policy
85 45 Sociology
86 23 Software Engineering
87 87 Soil Science
88 115 Solid Mechanics
89 120 Special Relativity
90 105 Statistics
91 55 Stellar Astronomy
92 47 Stereochemistry
93 75 Supramolecular Chemistry
94 76 Theoretical Computer Science
95 116 Thermodynamics
96 44 Toxicology
And we can add the category or categories we like,
fs_add_categories(id, c("Education", "Software Engineering"))
The file we have created remains saved as a draft until we publish it, either publicly or privately. Note that once a file is declared public, it cannot be made private or deleted. Let's release this dataset privately:
fs_make_private(id)
Response [http://api.figshare.com/v1/my_data/articles/105137/action/make_private]
Status: 200
Content-type: application/json; charset=UTF-8
{"success": "Article status changed to Private"}
We can now share the dataset with collaborators by way of the private url.
The quick and easy way
The rfigshare package will let you create a new figshare article with additional authors, tags, categories, etc in a single command usnig the fs_new_article function. The essential metadata title, description and type are required, but any other information we omit at this stage can be added later. If we set visibility to private or public, the article is published on figshare immediately.
id <- fs_new_article(title="A Test of rfigshare",
description="This is a test",
type="figure",
authors=c("Karthik Ram", "Scott Chamberlain"),
tags=c("ecology", "openscience"),
categories="Ecology",
links="http://ropensci.org",
files="figure/rfigshare.png",
visibility="private")
text_content() deprecated. Use parsed_content(x, as = 'parsed')
Your article has been created! Your id number is 105138
text_content() deprecated. Use parsed_content(x, as = 'parsed')
text_content() deprecated. Use parsed_content(x, as = 'parsed')
found ids for all authors
Examining Data on Figshare
We can view all available metadata of a figshare object. If the object is not public (e.g. draft or private), we have to add the mine=TRUE option
fs_details(id, mine=TRUE)
text_content() deprecated. Use parsed_content(x, as = 'parsed')
Article ID : 105138
Article type : figure
DOI :
Title : A Test of rfigshare
Description : This is a test
Shares :
Views :
Downloads :
Owner :
Authors : Carl Boettiger, Karthik Ram, Scott Chamberlain
Tags : openscience, ecology
Categories : Ecology
File names : rfigshare.png
Links : http://ropensci.org
You can see all of the files you have:
fs_browse(mine=TRUE)
text_content() deprecated. Use parsed_content(x, as = 'parsed')
[[1]]
[[1]]$article_id
[1] 105138
[[1]]$title
[1] "A Test of rfigshare"
[[1]]$defined_type
[1] "figure"
[[1]]$status
[1] "Private"
[[1]]$version
[1] 1
[[1]]$published_date
[1] "05:18, Dec 20, 2012"
[[1]]$description
[1] "This is a test"
[[1]]$description_nohtml
[1] "This is a test"
[[1]]$total_size
[1] "17.78 KB"
[[1]]$authors
[[1]]$authors[[1]]
[[1]]$authors[[1]]$first_name
[1] "Carl"
[[1]]$authors[[1]]$last_name
[1] "Boettiger"
[[1]]$authors[[1]]$id
[1] 96387
[[1]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[1]]$authors[[2]]
[[1]]$authors[[2]]$first_name
[1] "Karthik"
[[1]]$authors[[2]]$last_name
[1] "Ram"
[[1]]$authors[[2]]$id
[1] 97306
[[1]]$authors[[2]]$full_name
[1] "Karthik Ram"
[[1]]$authors[[3]]
[[1]]$authors[[3]]$first_name
[1] "Scott"
[[1]]$authors[[3]]$last_name
[1] "Chamberlain"
[[1]]$authors[[3]]$id
[1] 96554
[[1]]$authors[[3]]$full_name
[1] "Scott Chamberlain"
[[1]]$tags
[[1]]$tags[[1]]
[[1]]$tags[[1]]$id
[1] 47864
[[1]]$tags[[1]]$name
[1] "openscience"
[[1]]$tags[[2]]
[[1]]$tags[[2]]$id
[1] 11917
[[1]]$tags[[2]]$name
[1] "ecology"
[[1]]$categories
[[1]]$categories[[1]]
[[1]]$categories[[1]]$id
[1] 39
[[1]]$categories[[1]]$name
[1] "Ecology"
[[1]]$files
[[1]]$files[[1]]
[[1]]$files[[1]]$size
[1] "18 KB"
[[1]]$files[[1]]$id
[1] 233421
[[1]]$files[[1]]$mime_type
[1] "image/png"
[[1]]$files[[1]]$name
[1] "rfigshare.png"
[[1]]$links
[[1]]$links[[1]]
[[1]]$links[[1]]$link
[1] "http://ropensci.org"
[[1]]$links[[1]]$id
[1] 673
[[2]]
[[2]]$article_id
[1] 105137
[[2]]$title
[1] "Test title"
[[2]]$defined_type
[1] "dataset"
[[2]]$status
[1] "Private"
[[2]]$version
[1] 1
[[2]]$published_date
[1] "05:18, Dec 20, 2012"
[[2]]$description
[1] "description of test"
[[2]]$description_nohtml
[1] "description of test"
[[2]]$total_size
[1] "1.70 KB"
[[2]]$authors
[[2]]$authors[[1]]
[[2]]$authors[[1]]$first_name
[1] "Carl"
[[2]]$authors[[1]]$last_name
[1] "Boettiger"
[[2]]$authors[[1]]$id
[1] 96387
[[2]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[2]]$tags
[[2]]$tags[[1]]
[[2]]$tags[[1]]$id
[1] 15681
[[2]]$tags[[1]]$name
[1] "demo"
[[2]]$categories
[[2]]$categories[[1]]
[[2]]$categories[[1]]$id
[1] 23
[[2]]$categories[[1]]$name
[1] "Software Engineering"
[[2]]$categories[[2]]
[[2]]$categories[[2]]$id
[1] 129
[[2]]$categories[[2]]$name
[1] "Education"
[[2]]$files
[[2]]$files[[1]]
[[2]]$files[[1]]$size
[1] "2 KB"
[[2]]$files[[1]]$id
[1] 233420
[[2]]$files[[1]]$mime_type
[1] "text/plain"
[[2]]$files[[1]]$name
[1] "mtcars.csv"
[[2]]$links
list()
[[3]]
[[3]]$article_id
[1] 97653
[[3]]$title
[1] "RFigshare Tutorial"
[[3]]$defined_type
[1] "paper"
[[3]]$status
[1] "Public"
[[3]]$version
[1] 2
[[3]]$published_date
[1] "17:16, Nov 16, 2012"
[[3]]$description
[1] "<p>A tuturial on how to setup and post material to figshare from R</p>"
[[3]]$description_nohtml
[1] "A tuturial on how to setup and post material to figshare from R"
[[3]]$total_size
[1] "16.21 KB"
[[3]]$authors
[[3]]$authors[[1]]
[[3]]$authors[[1]]$first_name
[1] "Edmund"
[[3]]$authors[[1]]$last_name
[1] "Hart"
[[3]]$authors[[1]]$id
[1] 98137
[[3]]$authors[[1]]$full_name
[1] "Edmund Hart"
[[3]]$authors[[2]]
[[3]]$authors[[2]]$first_name
[1] "Carl"
[[3]]$authors[[2]]$last_name
[1] "Boettiger"
[[3]]$authors[[2]]$id
[1] 96387
[[3]]$authors[[2]]$full_name
[1] "Carl Boettiger"
[[3]]$tags
[[3]]$tags[[1]]
[[3]]$tags[[1]]$id
[1] 47864
[[3]]$tags[[1]]$name
[1] "openscience"
[[3]]$tags[[2]]
[[3]]$tags[[2]]$id
[1] 11917
[[3]]$tags[[2]]$name
[1] "ecology"
[[3]]$categories
[[3]]$categories[[1]]
[[3]]$categories[[1]]$id
[1] 39
[[3]]$categories[[1]]$name
[1] "Ecology"
[[3]]$files
[[3]]$files[[1]]
[[3]]$files[[1]]$download_url
[1] "http://files.figshare.com/223958/tutorial.md"
[[3]]$files[[1]]$size
[1] "17 KB"
[[3]]$files[[1]]$id
[1] 223958
[[3]]$files[[1]]$mime_type
[1] "text/plain"
[[3]]$files[[1]]$name
[1] "tutorial.md"
[[3]]$links
[[3]]$links[[1]]
[[3]]$links[[1]]$link
[1] "http://ropensci.org"
[[3]]$links[[1]]$id
[1] 673
[[4]]
[[4]]$article_id
[1] 97500
[[4]]$title
[1] "Exit Seminar to Center for Population Biology: Regime Shifts in Ecology and Evolution"
[[4]]$defined_type
[1] "presentation"
[[4]]$status
[1] "Public"
[[4]]$version
[1] 2
[[4]]$published_date
[1] "21:04, Nov 14, 2012"
[[4]]$description
[1] "<p>Slides from my exit seminar satisfying the completion requirements for a PhD in Population Biology at UC Davis, November 13, 2012.</p>"
[[4]]$description_nohtml
[1] "Slides from my exit seminar satisfying the completion requirements for a PhD in Population Biology at UC Davis, November 13, 2012."
[[4]]$total_size
[1] "23.37 MB"
[[4]]$authors
[[4]]$authors[[1]]
[[4]]$authors[[1]]$first_name
[1] "Carl"
[[4]]$authors[[1]]$last_name
[1] "Boettiger"
[[4]]$authors[[1]]$id
[1] 96387
[[4]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[4]]$tags
[[4]]$tags[[1]]
[[4]]$tags[[1]]$id
[1] 49296
[[4]]$tags[[1]]$name
[1] " comparative methods"
[[4]]$tags[[2]]
[[4]]$tags[[2]]$id
[1] 49295
[[4]]$tags[[2]]$name
[1] "seminar"
[[4]]$tags[[3]]
[[4]]$tags[[3]]$id
[1] 49294
[[4]]$tags[[3]]$name
[1] " presentation"
[[4]]$tags[[4]]
[[4]]$tags[[4]]$id
[1] 49293
[[4]]$tags[[4]]$name
[1] " labrids"
[[4]]$tags[[5]]
[[4]]$tags[[5]]$id
[1] 49036
[[4]]$tags[[5]]$name
[1] " warning singals"
[[4]]$tags[[6]]
[[4]]$tags[[6]]$id
[1] 47771
[[4]]$tags[[6]]$name
[1] " phylogenetics"
[[4]]$categories
[[4]]$categories[[1]]
[[4]]$categories[[1]]$id
[1] 24
[[4]]$categories[[1]]$name
[1] "Evolutionary biology"
[[4]]$categories[[2]]
[[4]]$categories[[2]]$id
[1] 39
[[4]]$categories[[2]]$name
[1] "Ecology"
[[4]]$files
[[4]]$files[[1]]
[[4]]$files[[1]]$download_url
[1] "http://files.figshare.com/177712/boettiger.pdf"
[[4]]$files[[1]]$size
[1] "23.37 MB"
[[4]]$files[[1]]$id
[1] 177712
[[4]]$files[[1]]$mime_type
[1] "application/pdf"
[[4]]$files[[1]]$name
[1] "boettiger.pdf"
[[4]]$links
list()
[[5]]
[[5]]$article_id
[1] 97279
[[5]]$title
[1] "Presentation at the Computational Science Graduate Fellowship Conference (2012): Regime shifts in Ecology & Evolution"
[[5]]$defined_type
[1] "presentation"
[[5]]$status
[1] "Public"
[[5]]$version
[1] 2
[[5]]$published_date
[1] "00:19, Nov 09, 2012"
[[5]]$description
[1] "<p>Slides from my talk at CSGF 2012 in Washington DC. </p>\n<p>A video recording of the talk is available here: <a href=\"http://www.youtube.com/watch?v=xwIIVdyKe4o\">http://www.youtube.com/watch?v=xwIIVdyKe4o</a></p>"
[[5]]$description_nohtml
[1] "Slides from my talk at CSGF 2012 in Washington DC.A video recording of the talk is available here:http://www.youtube.com/watch?v=xwIIVdyKe4o"
[[5]]$total_size
[1] "13.39 MB"
[[5]]$authors
[[5]]$authors[[1]]
[[5]]$authors[[1]]$first_name
[1] "Carl"
[[5]]$authors[[1]]$last_name
[1] "Boettiger"
[[5]]$authors[[1]]$id
[1] 96387
[[5]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[5]]$tags
[[5]]$tags[[1]]
[[5]]$tags[[1]]$id
[1] 49084
[[5]]$tags[[1]]$name
[1] " hpc"
[[5]]$tags[[2]]
[[5]]$tags[[2]]$id
[1] 49035
[[5]]$tags[[2]]$name
[1] "regime shifts"
[[5]]$tags[[3]]
[[5]]$tags[[3]]$id
[1] 276
[[5]]$tags[[3]]$name
[1] "phylogenetics"
[[5]]$tags[[4]]
[[5]]$tags[[4]]$id
[1] 277
[[5]]$tags[[4]]$name
[1] "comparative methods"
[[5]]$categories
[[5]]$categories[[1]]
[[5]]$categories[[1]]$id
[1] 106
[[5]]$categories[[1]]$name
[1] "Science Policy"
[[5]]$categories[[2]]
[[5]]$categories[[2]]$id
[1] 24
[[5]]$categories[[2]]$name
[1] "Evolutionary biology"
[[5]]$categories[[3]]
[[5]]$categories[[3]]$id
[1] 34
[[5]]$categories[[3]]$name
[1] "Environmental science"
[[5]]$categories[[4]]
[[5]]$categories[[4]]$id
[1] 39
[[5]]$categories[[4]]$name
[1] "Ecology"
[[5]]$categories[[5]]
[[5]]$categories[[5]]$id
[1] 125
[[5]]$categories[[5]]$name
[1] "Bioinformatics"
[[5]]$categories[[6]]
[[5]]$categories[[6]]$id
[1] 77
[[5]]$categories[[6]]$name
[1] "Applied Computer Science"
[[5]]$files
[[5]]$files[[1]]
[[5]]$files[[1]]$download_url
[1] "http://files.figshare.com/143048/boettiger.pdf"
[[5]]$files[[1]]$size
[1] "13.39 MB"
[[5]]$files[[1]]$id
[1] 143048
[[5]]$files[[1]]$mime_type
[1] "application/pdf"
[[5]]$files[[1]]$name
[1] "boettiger.pdf"
[[5]]$links
[[5]]$links[[1]]
[[5]]$links[[1]]$link
[1] "http://www.youtube.com/watch?v=xwIIVdyKe4o"
[[5]]$links[[1]]$id
[1] 1255
[[6]]
[[6]]$article_id
[1] 97218
[[6]]$title
[1] "Regime shifts in ecology and evolution (PhD Dissertation)"
[[6]]$defined_type
[1] "paper"
[[6]]$status
[1] "Public"
[[6]]$version
[1] 1
[[6]]$published_date
[1] "18:34, Nov 06, 2012"
[[6]]$description
[1] "<p>The most pressing issues of our time are all characterized by sudden regime shifts: the collapse of marine fisheries or stock-markets, the overthrow of governments, shifts in global climate. Regime shifts, or sudden transitions in dynamical behavior of a system, underly many important phenomena in ecological and evolutionary problems. How do they arise? How can we identify when a shift has occurred? Can we forecast these shifts? Here I address each of these central questions in the context of a particular system. First, I show how stochasticity in eco-evolutionary dynamics can give rise two different domains, or regimes, governing the behavior of evolutionary trajectories (Boettiger et al., 2010). In the next chapter, I turn to the question of identifying evolutionary shifts from data using phylogenetic trees and morphological trait data of extant species (Boettiger et al., 2012). In the last chapter, I adapt the approach of the previous section which allowed me to quantify the information available in a given data set that could detect a shift into an approach for detecting regime shifts in ecological time series data before the occur (Boettiger and Hastings, 2012).</p>\n<div> </div>"
[[6]]$description_nohtml
[1] "The most pressing issues of our time are all characterized by sudden regime shifts: the collapse of marine fisheries or stock-markets, the overthrow of governments, shifts in global climate. Regime shifts, or sudden transitions in dynamical behavior of a system, underly many important phenomena in ecological and evolutionary problems. How do they arise? How can we identify when a shift has occurred? Can we forecast these shifts? Here I address each of these central questions in the context of a particular system. First, I show how stochasticity in eco-evolutionary dynamics can give rise two different domains, or regimes, governing the behavior of evolutionary trajectories (Boettiger et al., 2010). In the next chapter, I turn to the question of identifying evolutionary shifts from data using phylogenetic trees and morphological trait data of extant species (Boettiger et al., 2012). In the last chapter, I adapt the approach of the previous section which allowed me to quantify the information available in a given data set that could detect a shift into an approach for detecting regime shifts in ecological time series data before the occur (Boettiger and Hastings, 2012)."
[[6]]$total_size
[1] "1.91 MB"
[[6]]$authors
[[6]]$authors[[1]]
[[6]]$authors[[1]]$first_name
[1] "Carl"
[[6]]$authors[[1]]$last_name
[1] "Boettiger"
[[6]]$authors[[1]]$id
[1] 96387
[[6]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[6]]$tags
[[6]]$tags[[1]]
[[6]]$tags[[1]]$id
[1] 49036
[[6]]$tags[[1]]$name
[1] " warning singals"
[[6]]$tags[[2]]
[[6]]$tags[[2]]$id
[1] 49035
[[6]]$tags[[2]]$name
[1] "regime shifts"
[[6]]$categories
[[6]]$categories[[1]]
[[6]]$categories[[1]]$id
[1] 24
[[6]]$categories[[1]]$name
[1] "Evolutionary biology"
[[6]]$categories[[2]]
[[6]]$categories[[2]]$id
[1] 39
[[6]]$categories[[2]]$name
[1] "Ecology"
[[6]]$files
[[6]]$files[[1]]
[[6]]$files[[1]]$download_url
[1] "http://files.figshare.com/142977/dissertation.pdf"
[[6]]$files[[1]]$size
[1] "1.91 MB"
[[6]]$files[[1]]$id
[1] 142977
[[6]]$files[[1]]$mime_type
[1] "application/pdf"
[[6]]$files[[1]]$name
[1] "dissertation.pdf"
[[6]]$links
[[6]]$links[[1]]
[[6]]$links[[1]]$link
[1] "http://carlboettiger.info"
[[6]]$links[[1]]$id
[1] 1235
[[7]]
[[7]]$article_id
[1] 96919
[[7]]$title
[1] "Lab Notebook, 2011"
[[7]]$defined_type
[1] "fileset"
[[7]]$status
[1] "Public"
[[7]]$version
[1] 1
[[7]]$published_date
[1] "22:44, Oct 29, 2012"
[[7]]$description
[1] "<p>Permanent archive of Carl Boettiger's open lab notebook entries for the year 2011 (<a href=\"http://www.carlboettiger.info/archives.html\">http://www.carlboettiger.info/archives.html</a>). Entries are archived in plain text UTF-8. Written in pandoc-flavored Markdown. Meets the goals of the Data Management Plan: <a href=\"http://www.carlboettiger.info/2012/10/09/data-management-plan.html\">http://www.carlboettiger.info/2012/10/09/data-management-plan.html</a></p>"
[[7]]$description_nohtml
[1] "Permanent archive of Carl Boettiger's open lab notebook entries for the year 2011 (http://www.carlboettiger.info/archives.htmlhttp://www.carlboettiger.info/2012/10/09/data-management-plan.html"
[[7]]$total_size
[1] "334.22 KB"
[[7]]$authors
[[7]]$authors[[1]]
[[7]]$authors[[1]]$first_name
[1] "Carl"
[[7]]$authors[[1]]$last_name
[1] "Boettiger"
[[7]]$authors[[1]]$id
[1] 96387
[[7]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[7]]$tags
[[7]]$tags[[1]]
[[7]]$tags[[1]]$id
[1] 48404
[[7]]$tags[[1]]$name
[1] "Ecology"
[[7]]$tags[[2]]
[[7]]$tags[[2]]$id
[1] 11917
[[7]]$tags[[2]]$name
[1] "ecology"
[[7]]$tags[[3]]
[[7]]$tags[[3]]$id
[1] 46723
[[7]]$tags[[3]]$name
[1] " open science"
[[7]]$tags[[4]]
[[7]]$tags[[4]]$id
[1] 47395
[[7]]$tags[[4]]$name
[1] " evolution"
[[7]]$categories
[[7]]$categories[[1]]
[[7]]$categories[[1]]$id
[1] 24
[[7]]$categories[[1]]$name
[1] "Evolutionary biology"
[[7]]$categories[[2]]
[[7]]$categories[[2]]$id
[1] 39
[[7]]$categories[[2]]$name
[1] "Ecology"
[[7]]$files
[[7]]$files[[1]]
[[7]]$files[[1]]$download_url
[1] "http://files.figshare.com/101976/2011.tar.gz"
[[7]]$files[[1]]$size
[1] "342 KB"
[[7]]$files[[1]]$id
[1] 101976
[[7]]$files[[1]]$mime_type
[1] "application/x-gzip"
[[7]]$files[[1]]$name
[1] "2011.tar.gz"
[[7]]$links
[[7]]$links[[1]]
[[7]]$links[[1]]$link
[1] "http://www.carlboettiger.info/2011"
[[7]]$links[[1]]$id
[1] 1148
[[8]]
[[8]]$article_id
[1] 96916
[[8]]$title
[1] "Lab Notebook, 2010"
[[8]]$defined_type
[1] "fileset"
[[8]]$status
[1] "Public"
[[8]]$version
[1] 1
[[8]]$published_date
[1] "22:39, Oct 29, 2012"
[[8]]$description
[1] "<p>Permanent archive of Carl Boettiger's open lab notebook entries for the year 2010 (<a href=\"http://www.carlboettiger.info/archives.html\">http://www.carlboettiger.info/archives.html</a>). Entries are archived in plain text UTF-8. Written in pandoc-flavored Markdown. Meets the goals of the Data Management Plan: <a href=\"http://www.carlboettiger.info/2012/10/09/data-management-plan.html\">http://www.carlboettiger.info/2012/10/09/data-management-plan.html</a></p>"
[[8]]$description_nohtml
[1] "Permanent archive of Carl Boettiger's open lab notebook entries for the year 2010 (http://www.carlboettiger.info/archives.htmlhttp://www.carlboettiger.info/2012/10/09/data-management-plan.html"
[[8]]$total_size
[1] "253.80 KB"
[[8]]$authors
[[8]]$authors[[1]]
[[8]]$authors[[1]]$first_name
[1] "Carl"
[[8]]$authors[[1]]$last_name
[1] "Boettiger"
[[8]]$authors[[1]]$id
[1] 96387
[[8]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[8]]$tags
[[8]]$tags[[1]]
[[8]]$tags[[1]]$id
[1] 48404
[[8]]$tags[[1]]$name
[1] "Ecology"
[[8]]$tags[[2]]
[[8]]$tags[[2]]$id
[1] 11917
[[8]]$tags[[2]]$name
[1] "ecology"
[[8]]$tags[[3]]
[[8]]$tags[[3]]$id
[1] 46723
[[8]]$tags[[3]]$name
[1] " open science"
[[8]]$tags[[4]]
[[8]]$tags[[4]]$id
[1] 47395
[[8]]$tags[[4]]$name
[1] " evolution"
[[8]]$categories
[[8]]$categories[[1]]
[[8]]$categories[[1]]$id
[1] 24
[[8]]$categories[[1]]$name
[1] "Evolutionary biology"
[[8]]$categories[[2]]
[[8]]$categories[[2]]$id
[1] 39
[[8]]$categories[[2]]$name
[1] "Ecology"
[[8]]$files
[[8]]$files[[1]]
[[8]]$files[[1]]$download_url
[1] "http://files.figshare.com/101975/2010.tar.gz"
[[8]]$files[[1]]$size
[1] "260 KB"
[[8]]$files[[1]]$id
[1] 101975
[[8]]$files[[1]]$mime_type
[1] "application/x-gzip"
[[8]]$files[[1]]$name
[1] "2010.tar.gz"
[[8]]$links
list()
[[9]]
[[9]]$article_id
[1] 95839
[[9]]$title
[1] "R code for the function fs_create "
[[9]]$defined_type
[1] "dataset"
[[9]]$status
[1] "Public"
[[9]]$version
[1] 1
[[9]]$published_date
[1] "19:48, Sep 13, 2012"
[[9]]$description
[1] "<p>An R implementation of the figshare API function to create a new article. Look Ma, I can share nice code on figshare!</p>\n<p> </p>\n<p>This function is part of the <a href=\"https://github.com/ropensci/rfigshare\">rfigshare</a> package by the <a href=\"http://ropensci.org\">rOpenSci</a> project. </p>"
[[9]]$description_nohtml
[1] "An R implementation of the figshare API function to create a new article. Look Ma, I can share nice code on figshare!This function is part of therfigsharerOpenSci"
[[9]]$total_size
[1] "2.04 KB"
[[9]]$authors
[[9]]$authors[[1]]
[[9]]$authors[[1]]$first_name
[1] "Carl"
[[9]]$authors[[1]]$last_name
[1] "Boettiger"
[[9]]$authors[[1]]$id
[1] 96387
[[9]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[9]]$tags
[[9]]$tags[[1]]
[[9]]$tags[[1]]$id
[1] 47906
[[9]]$tags[[1]]$name
[1] " code"
[[9]]$tags[[2]]
[[9]]$tags[[2]]$id
[1] 47624
[[9]]$tags[[2]]$name
[1] "R"
[[9]]$tags[[3]]
[[9]]$tags[[3]]$id
[1] 42046
[[9]]$tags[[3]]$name
[1] "r"
[[9]]$categories
[[9]]$categories[[1]]
[[9]]$categories[[1]]$id
[1] 77
[[9]]$categories[[1]]$name
[1] "Applied Computer Science"
[[9]]$files
[[9]]$files[[1]]
[[9]]$files[[1]]$download_url
[1] "http://files.figshare.com/98377/fs_create.R"
[[9]]$files[[1]]$size
[1] "2 KB"
[[9]]$files[[1]]$id
[1] 98377
[[9]]$files[[1]]$mime_type
[1] "text/plain"
[[9]]$files[[1]]$name
[1] "fs_create.R"
[[9]]$links
list()
[[10]]
[[10]]$article_id
[1] 138
[[10]]$title
[1] "Labrid adaptive peaks"
[[10]]$defined_type
[1] "figure"
[[10]]$status
[1] "Public"
[[10]]$version
[1] 2
[[10]]$published_date
[1] "13:45, Dec 30, 2011"
[[10]]$description
[1] "<p>Described in the notebook: http://openwetware.org/wiki/User:Carl_Boettiger/Notebook/Comparative_Phylogenetics/2010/03/12</p>"
[[10]]$description_nohtml
[1] "Described in the notebook: http://openwetware.org/wiki/User:Carl_Boettiger/Notebook/Comparative_Phylogenetics/2010/03/12"
[[10]]$total_size
[1] "29.71 KB"
[[10]]$authors
[[10]]$authors[[1]]
[[10]]$authors[[1]]$first_name
[1] "Carl"
[[10]]$authors[[1]]$last_name
[1] "Boettiger"
[[10]]$authors[[1]]$id
[1] 96387
[[10]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[10]]$tags
[[10]]$tags[[1]]
[[10]]$tags[[1]]$id
[1] 277
[[10]]$tags[[1]]$name
[1] "comparative methods"
[[10]]$tags[[2]]
[[10]]$tags[[2]]$id
[1] 276
[[10]]$tags[[2]]$name
[1] "phylogenetics"
[[10]]$tags[[3]]
[[10]]$tags[[3]]$id
[1] 275
[[10]]$tags[[3]]$name
[1] "fins"
[[10]]$tags[[4]]
[[10]]$tags[[4]]$id
[1] 274
[[10]]$tags[[4]]$name
[1] "labrids"
[[10]]$categories
[[10]]$categories[[1]]
[[10]]$categories[[1]]$id
[1] 24
[[10]]$categories[[1]]$name
[1] "Evolutionary biology"
[[10]]$categories[[2]]
[[10]]$categories[[2]]$id
[1] 39
[[10]]$categories[[2]]$name
[1] "Ecology"
[[10]]$files
[[10]]$files[[1]]
[[10]]$files[[1]]$download_url
[1] "http://files.figshare.com/137/Labrid_fins.png"
[[10]]$files[[1]]$size
[1] "30 KB"
[[10]]$files[[1]]$id
[1] 137
[[10]]$files[[1]]$mime_type
[1] "image/png"
[[10]]$files[[1]]$name
[1] "Labrid_fins.png"
[[10]]$links
list()
Note that we can easily grab the ids with the wrapper function fs_ids:
fs_ids(all_mine)
[1] 105136 105135 97653 97500 97279 97218 96919 96916 95839 138
We can delete unwanted files that are not public with fs_delete:
fs_delete(id)
Try the rfigshare package in your browser
Any scripts or data that you put into this service are public.
rfigshare documentation built on May 10, 2022, 1:05 a.m.
title: rfigshare - Share and explore figures, data, and publications on FigShare using R

Obtaining your API keys
There is a nice video introduction to creating applications for the API on the figshare blog. The following tutorial provides a simple walkthrough of how to go about getting your figshare API keys set up so that you can use the rfigshare package.
Create a user account on FigShare and log in. From your homepage, select "Applications" from the drop-down menu,

Create a new application:

Enter in the following information:

Then navigate over to the permissions tab. To get the most out of rfigshare you'll want to enable all permissions:

Save the new settings, and then open the application again (View/Edit menu) and click on the "Access Codes" tab.

Record each if the keys into R as follows. You might want to put this bit of R code into your .Rprofile to avoid entering it each time in the future:
options(FigshareKey = "qMDabXXXXXXXXXXXXXXXXX")
options(FigsharePrivateKey = "zQXXXXXXXXXXXXXXXXXXXX")
options(FigshareToken = "SrpxabQXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX")
options(FigsharePrivateToken = "yqXXXXXXXXXXXXXXXXXXXX")
Installing the R package
Now that we have the API credentials in place, actually using rfigshare is pretty easy. Install the latest version of package directly from Github using:
require(devtools)
install_github("rfigshare", "ropensci")
Using rfigshare
Try authenticating with your credentials:
require(rfigshare)
fs_auth()
Try a search for an author:
fs_author_search("Boettiger")
text_content() deprecated. Use parsed_content(x, as = 'parsed')
[[1]]
[[1]]$id
[1] "96387"
[[1]]$fname
[1] "Carl"
[[1]]$lname
[1] "Boettiger"
[[1]]$full_name
[1] "Carl Boettiger"
[[1]]$job_title
[1] ""
[[1]]$description
[1] ""
[[1]]$facebook
[1] ""
[[1]]$twitter
[1] ""
[[1]]$active
[1] 1
Try creating your own content:
id <- fs_create("Test title", "description of test", "dataset")
text_content() deprecated. Use parsed_content(x, as = 'parsed')
Your article has been created! Your id number is 105137
This creates an article with the essential metadata information. We can now upload the dataset to this newly created figshare object using fs_upload. For the purposes of this example, we'll just upload one of R's built-in datasets:
data(mtcars)
write.csv(mtcars, "mtcars.csv")
fs_upload(id, "mtcars.csv")
Not that we must pass the id number of our the newly created figshare object as the first argument. Similar functions let us add additional authors, tags, categories, and links, e.g.
fs_add_tags(id, "demo")
Minimal metadata includes title, description, type, and at least one tag and one category. We can add categories using either the category id or the name of the category, but it must be one of the preset categories available. We can ask the API for a list of all the categories:
fs_category_list()
And we can add the category or categories we like,
fs_add_categories(id, c("Education", "Software Engineering"))
The file we have created remains saved as a draft until we publish it, either publicly or privately. Note that once a file is declared public, it cannot be made private or deleted. Let's release this dataset privately:
fs_make_private(id)
Response [http://api.figshare.com/v1/my_data/articles/105137/action/make_private]
Status: 200
Content-type: application/json; charset=UTF-8
{"success": "Article status changed to Private"}
We can now share the dataset with collaborators by way of the private url.
The quick and easy way
The rfigshare package will let you create a new figshare article with additional authors, tags, categories, etc in a single command usnig the fs_new_article function. The essential metadata title, description and type are required, but any other information we omit at this stage can be added later. If we set visibility to private or public, the article is published on figshare immediately.
id <- fs_new_article(title="A Test of rfigshare",
description="This is a test",
type="figure",
authors=c("Karthik Ram", "Scott Chamberlain"),
tags=c("ecology", "openscience"),
categories="Ecology",
links="http://ropensci.org",
files="figure/rfigshare.png",
visibility="private")
text_content() deprecated. Use parsed_content(x, as = 'parsed')
Your article has been created! Your id number is 105138
text_content() deprecated. Use parsed_content(x, as = 'parsed')
text_content() deprecated. Use parsed_content(x, as = 'parsed')
found ids for all authors
Examining Data on Figshare
We can view all available metadata of a figshare object. If the object is not public (e.g. draft or private), we have to add the mine=TRUE option
fs_details(id, mine=TRUE)
text_content() deprecated. Use parsed_content(x, as = 'parsed')
Article ID : 105138
Article type : figure
DOI :
Title : A Test of rfigshare
Description : This is a test
Shares :
Views :
Downloads :
Owner :
Authors : Carl Boettiger, Karthik Ram, Scott Chamberlain
Tags : openscience, ecology
Categories : Ecology
File names : rfigshare.png
Links : http://ropensci.org
You can see all of the files you have:
fs_browse(mine=TRUE)
text_content() deprecated. Use parsed_content(x, as = 'parsed')
[[1]]
[[1]]$article_id
[1] 105138
[[1]]$title
[1] "A Test of rfigshare"
[[1]]$defined_type
[1] "figure"
[[1]]$status
[1] "Private"
[[1]]$version
[1] 1
[[1]]$published_date
[1] "05:18, Dec 20, 2012"
[[1]]$description
[1] "This is a test"
[[1]]$description_nohtml
[1] "This is a test"
[[1]]$total_size
[1] "17.78 KB"
[[1]]$authors
[[1]]$authors[[1]]
[[1]]$authors[[1]]$first_name
[1] "Carl"
[[1]]$authors[[1]]$last_name
[1] "Boettiger"
[[1]]$authors[[1]]$id
[1] 96387
[[1]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[1]]$authors[[2]]
[[1]]$authors[[2]]$first_name
[1] "Karthik"
[[1]]$authors[[2]]$last_name
[1] "Ram"
[[1]]$authors[[2]]$id
[1] 97306
[[1]]$authors[[2]]$full_name
[1] "Karthik Ram"
[[1]]$authors[[3]]
[[1]]$authors[[3]]$first_name
[1] "Scott"
[[1]]$authors[[3]]$last_name
[1] "Chamberlain"
[[1]]$authors[[3]]$id
[1] 96554
[[1]]$authors[[3]]$full_name
[1] "Scott Chamberlain"
[[1]]$tags
[[1]]$tags[[1]]
[[1]]$tags[[1]]$id
[1] 47864
[[1]]$tags[[1]]$name
[1] "openscience"
[[1]]$tags[[2]]
[[1]]$tags[[2]]$id
[1] 11917
[[1]]$tags[[2]]$name
[1] "ecology"
[[1]]$categories
[[1]]$categories[[1]]
[[1]]$categories[[1]]$id
[1] 39
[[1]]$categories[[1]]$name
[1] "Ecology"
[[1]]$files
[[1]]$files[[1]]
[[1]]$files[[1]]$size
[1] "18 KB"
[[1]]$files[[1]]$id
[1] 233421
[[1]]$files[[1]]$mime_type
[1] "image/png"
[[1]]$files[[1]]$name
[1] "rfigshare.png"
[[1]]$links
[[1]]$links[[1]]
[[1]]$links[[1]]$link
[1] "http://ropensci.org"
[[1]]$links[[1]]$id
[1] 673
[[2]]
[[2]]$article_id
[1] 105137
[[2]]$title
[1] "Test title"
[[2]]$defined_type
[1] "dataset"
[[2]]$status
[1] "Private"
[[2]]$version
[1] 1
[[2]]$published_date
[1] "05:18, Dec 20, 2012"
[[2]]$description
[1] "description of test"
[[2]]$description_nohtml
[1] "description of test"
[[2]]$total_size
[1] "1.70 KB"
[[2]]$authors
[[2]]$authors[[1]]
[[2]]$authors[[1]]$first_name
[1] "Carl"
[[2]]$authors[[1]]$last_name
[1] "Boettiger"
[[2]]$authors[[1]]$id
[1] 96387
[[2]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[2]]$tags
[[2]]$tags[[1]]
[[2]]$tags[[1]]$id
[1] 15681
[[2]]$tags[[1]]$name
[1] "demo"
[[2]]$categories
[[2]]$categories[[1]]
[[2]]$categories[[1]]$id
[1] 23
[[2]]$categories[[1]]$name
[1] "Software Engineering"
[[2]]$categories[[2]]
[[2]]$categories[[2]]$id
[1] 129
[[2]]$categories[[2]]$name
[1] "Education"
[[2]]$files
[[2]]$files[[1]]
[[2]]$files[[1]]$size
[1] "2 KB"
[[2]]$files[[1]]$id
[1] 233420
[[2]]$files[[1]]$mime_type
[1] "text/plain"
[[2]]$files[[1]]$name
[1] "mtcars.csv"
[[2]]$links
list()
[[3]]
[[3]]$article_id
[1] 97653
[[3]]$title
[1] "RFigshare Tutorial"
[[3]]$defined_type
[1] "paper"
[[3]]$status
[1] "Public"
[[3]]$version
[1] 2
[[3]]$published_date
[1] "17:16, Nov 16, 2012"
[[3]]$description
[1] "<p>A tuturial on how to setup and post material to figshare from R</p>"
[[3]]$description_nohtml
[1] "A tuturial on how to setup and post material to figshare from R"
[[3]]$total_size
[1] "16.21 KB"
[[3]]$authors
[[3]]$authors[[1]]
[[3]]$authors[[1]]$first_name
[1] "Edmund"
[[3]]$authors[[1]]$last_name
[1] "Hart"
[[3]]$authors[[1]]$id
[1] 98137
[[3]]$authors[[1]]$full_name
[1] "Edmund Hart"
[[3]]$authors[[2]]
[[3]]$authors[[2]]$first_name
[1] "Carl"
[[3]]$authors[[2]]$last_name
[1] "Boettiger"
[[3]]$authors[[2]]$id
[1] 96387
[[3]]$authors[[2]]$full_name
[1] "Carl Boettiger"
[[3]]$tags
[[3]]$tags[[1]]
[[3]]$tags[[1]]$id
[1] 47864
[[3]]$tags[[1]]$name
[1] "openscience"
[[3]]$tags[[2]]
[[3]]$tags[[2]]$id
[1] 11917
[[3]]$tags[[2]]$name
[1] "ecology"
[[3]]$categories
[[3]]$categories[[1]]
[[3]]$categories[[1]]$id
[1] 39
[[3]]$categories[[1]]$name
[1] "Ecology"
[[3]]$files
[[3]]$files[[1]]
[[3]]$files[[1]]$download_url
[1] "http://files.figshare.com/223958/tutorial.md"
[[3]]$files[[1]]$size
[1] "17 KB"
[[3]]$files[[1]]$id
[1] 223958
[[3]]$files[[1]]$mime_type
[1] "text/plain"
[[3]]$files[[1]]$name
[1] "tutorial.md"
[[3]]$links
[[3]]$links[[1]]
[[3]]$links[[1]]$link
[1] "http://ropensci.org"
[[3]]$links[[1]]$id
[1] 673
[[4]]
[[4]]$article_id
[1] 97500
[[4]]$title
[1] "Exit Seminar to Center for Population Biology: Regime Shifts in Ecology and Evolution"
[[4]]$defined_type
[1] "presentation"
[[4]]$status
[1] "Public"
[[4]]$version
[1] 2
[[4]]$published_date
[1] "21:04, Nov 14, 2012"
[[4]]$description
[1] "<p>Slides from my exit seminar satisfying the completion requirements for a PhD in Population Biology at UC Davis, November 13, 2012.</p>"
[[4]]$description_nohtml
[1] "Slides from my exit seminar satisfying the completion requirements for a PhD in Population Biology at UC Davis, November 13, 2012."
[[4]]$total_size
[1] "23.37 MB"
[[4]]$authors
[[4]]$authors[[1]]
[[4]]$authors[[1]]$first_name
[1] "Carl"
[[4]]$authors[[1]]$last_name
[1] "Boettiger"
[[4]]$authors[[1]]$id
[1] 96387
[[4]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[4]]$tags
[[4]]$tags[[1]]
[[4]]$tags[[1]]$id
[1] 49296
[[4]]$tags[[1]]$name
[1] " comparative methods"
[[4]]$tags[[2]]
[[4]]$tags[[2]]$id
[1] 49295
[[4]]$tags[[2]]$name
[1] "seminar"
[[4]]$tags[[3]]
[[4]]$tags[[3]]$id
[1] 49294
[[4]]$tags[[3]]$name
[1] " presentation"
[[4]]$tags[[4]]
[[4]]$tags[[4]]$id
[1] 49293
[[4]]$tags[[4]]$name
[1] " labrids"
[[4]]$tags[[5]]
[[4]]$tags[[5]]$id
[1] 49036
[[4]]$tags[[5]]$name
[1] " warning singals"
[[4]]$tags[[6]]
[[4]]$tags[[6]]$id
[1] 47771
[[4]]$tags[[6]]$name
[1] " phylogenetics"
[[4]]$categories
[[4]]$categories[[1]]
[[4]]$categories[[1]]$id
[1] 24
[[4]]$categories[[1]]$name
[1] "Evolutionary biology"
[[4]]$categories[[2]]
[[4]]$categories[[2]]$id
[1] 39
[[4]]$categories[[2]]$name
[1] "Ecology"
[[4]]$files
[[4]]$files[[1]]
[[4]]$files[[1]]$download_url
[1] "http://files.figshare.com/177712/boettiger.pdf"
[[4]]$files[[1]]$size
[1] "23.37 MB"
[[4]]$files[[1]]$id
[1] 177712
[[4]]$files[[1]]$mime_type
[1] "application/pdf"
[[4]]$files[[1]]$name
[1] "boettiger.pdf"
[[4]]$links
list()
[[5]]
[[5]]$article_id
[1] 97279
[[5]]$title
[1] "Presentation at the Computational Science Graduate Fellowship Conference (2012): Regime shifts in Ecology & Evolution"
[[5]]$defined_type
[1] "presentation"
[[5]]$status
[1] "Public"
[[5]]$version
[1] 2
[[5]]$published_date
[1] "00:19, Nov 09, 2012"
[[5]]$description
[1] "<p>Slides from my talk at CSGF 2012 in Washington DC. </p>\n<p>A video recording of the talk is available here: <a href=\"http://www.youtube.com/watch?v=xwIIVdyKe4o\">http://www.youtube.com/watch?v=xwIIVdyKe4o</a></p>"
[[5]]$description_nohtml
[1] "Slides from my talk at CSGF 2012 in Washington DC.A video recording of the talk is available here:http://www.youtube.com/watch?v=xwIIVdyKe4o"
[[5]]$total_size
[1] "13.39 MB"
[[5]]$authors
[[5]]$authors[[1]]
[[5]]$authors[[1]]$first_name
[1] "Carl"
[[5]]$authors[[1]]$last_name
[1] "Boettiger"
[[5]]$authors[[1]]$id
[1] 96387
[[5]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[5]]$tags
[[5]]$tags[[1]]
[[5]]$tags[[1]]$id
[1] 49084
[[5]]$tags[[1]]$name
[1] " hpc"
[[5]]$tags[[2]]
[[5]]$tags[[2]]$id
[1] 49035
[[5]]$tags[[2]]$name
[1] "regime shifts"
[[5]]$tags[[3]]
[[5]]$tags[[3]]$id
[1] 276
[[5]]$tags[[3]]$name
[1] "phylogenetics"
[[5]]$tags[[4]]
[[5]]$tags[[4]]$id
[1] 277
[[5]]$tags[[4]]$name
[1] "comparative methods"
[[5]]$categories
[[5]]$categories[[1]]
[[5]]$categories[[1]]$id
[1] 106
[[5]]$categories[[1]]$name
[1] "Science Policy"
[[5]]$categories[[2]]
[[5]]$categories[[2]]$id
[1] 24
[[5]]$categories[[2]]$name
[1] "Evolutionary biology"
[[5]]$categories[[3]]
[[5]]$categories[[3]]$id
[1] 34
[[5]]$categories[[3]]$name
[1] "Environmental science"
[[5]]$categories[[4]]
[[5]]$categories[[4]]$id
[1] 39
[[5]]$categories[[4]]$name
[1] "Ecology"
[[5]]$categories[[5]]
[[5]]$categories[[5]]$id
[1] 125
[[5]]$categories[[5]]$name
[1] "Bioinformatics"
[[5]]$categories[[6]]
[[5]]$categories[[6]]$id
[1] 77
[[5]]$categories[[6]]$name
[1] "Applied Computer Science"
[[5]]$files
[[5]]$files[[1]]
[[5]]$files[[1]]$download_url
[1] "http://files.figshare.com/143048/boettiger.pdf"
[[5]]$files[[1]]$size
[1] "13.39 MB"
[[5]]$files[[1]]$id
[1] 143048
[[5]]$files[[1]]$mime_type
[1] "application/pdf"
[[5]]$files[[1]]$name
[1] "boettiger.pdf"
[[5]]$links
[[5]]$links[[1]]
[[5]]$links[[1]]$link
[1] "http://www.youtube.com/watch?v=xwIIVdyKe4o"
[[5]]$links[[1]]$id
[1] 1255
[[6]]
[[6]]$article_id
[1] 97218
[[6]]$title
[1] "Regime shifts in ecology and evolution (PhD Dissertation)"
[[6]]$defined_type
[1] "paper"
[[6]]$status
[1] "Public"
[[6]]$version
[1] 1
[[6]]$published_date
[1] "18:34, Nov 06, 2012"
[[6]]$description
[1] "<p>The most pressing issues of our time are all characterized by sudden regime shifts: the collapse of marine fisheries or stock-markets, the overthrow of governments, shifts in global climate. Regime shifts, or sudden transitions in dynamical behavior of a system, underly many important phenomena in ecological and evolutionary problems. How do they arise? How can we identify when a shift has occurred? Can we forecast these shifts? Here I address each of these central questions in the context of a particular system. First, I show how stochasticity in eco-evolutionary dynamics can give rise two different domains, or regimes, governing the behavior of evolutionary trajectories (Boettiger et al., 2010). In the next chapter, I turn to the question of identifying evolutionary shifts from data using phylogenetic trees and morphological trait data of extant species (Boettiger et al., 2012). In the last chapter, I adapt the approach of the previous section which allowed me to quantify the information available in a given data set that could detect a shift into an approach for detecting regime shifts in ecological time series data before the occur (Boettiger and Hastings, 2012).</p>\n<div> </div>"
[[6]]$description_nohtml
[1] "The most pressing issues of our time are all characterized by sudden regime shifts: the collapse of marine fisheries or stock-markets, the overthrow of governments, shifts in global climate. Regime shifts, or sudden transitions in dynamical behavior of a system, underly many important phenomena in ecological and evolutionary problems. How do they arise? How can we identify when a shift has occurred? Can we forecast these shifts? Here I address each of these central questions in the context of a particular system. First, I show how stochasticity in eco-evolutionary dynamics can give rise two different domains, or regimes, governing the behavior of evolutionary trajectories (Boettiger et al., 2010). In the next chapter, I turn to the question of identifying evolutionary shifts from data using phylogenetic trees and morphological trait data of extant species (Boettiger et al., 2012). In the last chapter, I adapt the approach of the previous section which allowed me to quantify the information available in a given data set that could detect a shift into an approach for detecting regime shifts in ecological time series data before the occur (Boettiger and Hastings, 2012)."
[[6]]$total_size
[1] "1.91 MB"
[[6]]$authors
[[6]]$authors[[1]]
[[6]]$authors[[1]]$first_name
[1] "Carl"
[[6]]$authors[[1]]$last_name
[1] "Boettiger"
[[6]]$authors[[1]]$id
[1] 96387
[[6]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[6]]$tags
[[6]]$tags[[1]]
[[6]]$tags[[1]]$id
[1] 49036
[[6]]$tags[[1]]$name
[1] " warning singals"
[[6]]$tags[[2]]
[[6]]$tags[[2]]$id
[1] 49035
[[6]]$tags[[2]]$name
[1] "regime shifts"
[[6]]$categories
[[6]]$categories[[1]]
[[6]]$categories[[1]]$id
[1] 24
[[6]]$categories[[1]]$name
[1] "Evolutionary biology"
[[6]]$categories[[2]]
[[6]]$categories[[2]]$id
[1] 39
[[6]]$categories[[2]]$name
[1] "Ecology"
[[6]]$files
[[6]]$files[[1]]
[[6]]$files[[1]]$download_url
[1] "http://files.figshare.com/142977/dissertation.pdf"
[[6]]$files[[1]]$size
[1] "1.91 MB"
[[6]]$files[[1]]$id
[1] 142977
[[6]]$files[[1]]$mime_type
[1] "application/pdf"
[[6]]$files[[1]]$name
[1] "dissertation.pdf"
[[6]]$links
[[6]]$links[[1]]
[[6]]$links[[1]]$link
[1] "http://carlboettiger.info"
[[6]]$links[[1]]$id
[1] 1235
[[7]]
[[7]]$article_id
[1] 96919
[[7]]$title
[1] "Lab Notebook, 2011"
[[7]]$defined_type
[1] "fileset"
[[7]]$status
[1] "Public"
[[7]]$version
[1] 1
[[7]]$published_date
[1] "22:44, Oct 29, 2012"
[[7]]$description
[1] "<p>Permanent archive of Carl Boettiger's open lab notebook entries for the year 2011 (<a href=\"http://www.carlboettiger.info/archives.html\">http://www.carlboettiger.info/archives.html</a>). Entries are archived in plain text UTF-8. Written in pandoc-flavored Markdown. Meets the goals of the Data Management Plan: <a href=\"http://www.carlboettiger.info/2012/10/09/data-management-plan.html\">http://www.carlboettiger.info/2012/10/09/data-management-plan.html</a></p>"
[[7]]$description_nohtml
[1] "Permanent archive of Carl Boettiger's open lab notebook entries for the year 2011 (http://www.carlboettiger.info/archives.htmlhttp://www.carlboettiger.info/2012/10/09/data-management-plan.html"
[[7]]$total_size
[1] "334.22 KB"
[[7]]$authors
[[7]]$authors[[1]]
[[7]]$authors[[1]]$first_name
[1] "Carl"
[[7]]$authors[[1]]$last_name
[1] "Boettiger"
[[7]]$authors[[1]]$id
[1] 96387
[[7]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[7]]$tags
[[7]]$tags[[1]]
[[7]]$tags[[1]]$id
[1] 48404
[[7]]$tags[[1]]$name
[1] "Ecology"
[[7]]$tags[[2]]
[[7]]$tags[[2]]$id
[1] 11917
[[7]]$tags[[2]]$name
[1] "ecology"
[[7]]$tags[[3]]
[[7]]$tags[[3]]$id
[1] 46723
[[7]]$tags[[3]]$name
[1] " open science"
[[7]]$tags[[4]]
[[7]]$tags[[4]]$id
[1] 47395
[[7]]$tags[[4]]$name
[1] " evolution"
[[7]]$categories
[[7]]$categories[[1]]
[[7]]$categories[[1]]$id
[1] 24
[[7]]$categories[[1]]$name
[1] "Evolutionary biology"
[[7]]$categories[[2]]
[[7]]$categories[[2]]$id
[1] 39
[[7]]$categories[[2]]$name
[1] "Ecology"
[[7]]$files
[[7]]$files[[1]]
[[7]]$files[[1]]$download_url
[1] "http://files.figshare.com/101976/2011.tar.gz"
[[7]]$files[[1]]$size
[1] "342 KB"
[[7]]$files[[1]]$id
[1] 101976
[[7]]$files[[1]]$mime_type
[1] "application/x-gzip"
[[7]]$files[[1]]$name
[1] "2011.tar.gz"
[[7]]$links
[[7]]$links[[1]]
[[7]]$links[[1]]$link
[1] "http://www.carlboettiger.info/2011"
[[7]]$links[[1]]$id
[1] 1148
[[8]]
[[8]]$article_id
[1] 96916
[[8]]$title
[1] "Lab Notebook, 2010"
[[8]]$defined_type
[1] "fileset"
[[8]]$status
[1] "Public"
[[8]]$version
[1] 1
[[8]]$published_date
[1] "22:39, Oct 29, 2012"
[[8]]$description
[1] "<p>Permanent archive of Carl Boettiger's open lab notebook entries for the year 2010 (<a href=\"http://www.carlboettiger.info/archives.html\">http://www.carlboettiger.info/archives.html</a>). Entries are archived in plain text UTF-8. Written in pandoc-flavored Markdown. Meets the goals of the Data Management Plan: <a href=\"http://www.carlboettiger.info/2012/10/09/data-management-plan.html\">http://www.carlboettiger.info/2012/10/09/data-management-plan.html</a></p>"
[[8]]$description_nohtml
[1] "Permanent archive of Carl Boettiger's open lab notebook entries for the year 2010 (http://www.carlboettiger.info/archives.htmlhttp://www.carlboettiger.info/2012/10/09/data-management-plan.html"
[[8]]$total_size
[1] "253.80 KB"
[[8]]$authors
[[8]]$authors[[1]]
[[8]]$authors[[1]]$first_name
[1] "Carl"
[[8]]$authors[[1]]$last_name
[1] "Boettiger"
[[8]]$authors[[1]]$id
[1] 96387
[[8]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[8]]$tags
[[8]]$tags[[1]]
[[8]]$tags[[1]]$id
[1] 48404
[[8]]$tags[[1]]$name
[1] "Ecology"
[[8]]$tags[[2]]
[[8]]$tags[[2]]$id
[1] 11917
[[8]]$tags[[2]]$name
[1] "ecology"
[[8]]$tags[[3]]
[[8]]$tags[[3]]$id
[1] 46723
[[8]]$tags[[3]]$name
[1] " open science"
[[8]]$tags[[4]]
[[8]]$tags[[4]]$id
[1] 47395
[[8]]$tags[[4]]$name
[1] " evolution"
[[8]]$categories
[[8]]$categories[[1]]
[[8]]$categories[[1]]$id
[1] 24
[[8]]$categories[[1]]$name
[1] "Evolutionary biology"
[[8]]$categories[[2]]
[[8]]$categories[[2]]$id
[1] 39
[[8]]$categories[[2]]$name
[1] "Ecology"
[[8]]$files
[[8]]$files[[1]]
[[8]]$files[[1]]$download_url
[1] "http://files.figshare.com/101975/2010.tar.gz"
[[8]]$files[[1]]$size
[1] "260 KB"
[[8]]$files[[1]]$id
[1] 101975
[[8]]$files[[1]]$mime_type
[1] "application/x-gzip"
[[8]]$files[[1]]$name
[1] "2010.tar.gz"
[[8]]$links
list()
[[9]]
[[9]]$article_id
[1] 95839
[[9]]$title
[1] "R code for the function fs_create "
[[9]]$defined_type
[1] "dataset"
[[9]]$status
[1] "Public"
[[9]]$version
[1] 1
[[9]]$published_date
[1] "19:48, Sep 13, 2012"
[[9]]$description
[1] "<p>An R implementation of the figshare API function to create a new article. Look Ma, I can share nice code on figshare!</p>\n<p> </p>\n<p>This function is part of the <a href=\"https://github.com/ropensci/rfigshare\">rfigshare</a> package by the <a href=\"http://ropensci.org\">rOpenSci</a> project. </p>"
[[9]]$description_nohtml
[1] "An R implementation of the figshare API function to create a new article. Look Ma, I can share nice code on figshare!This function is part of therfigsharerOpenSci"
[[9]]$total_size
[1] "2.04 KB"
[[9]]$authors
[[9]]$authors[[1]]
[[9]]$authors[[1]]$first_name
[1] "Carl"
[[9]]$authors[[1]]$last_name
[1] "Boettiger"
[[9]]$authors[[1]]$id
[1] 96387
[[9]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[9]]$tags
[[9]]$tags[[1]]
[[9]]$tags[[1]]$id
[1] 47906
[[9]]$tags[[1]]$name
[1] " code"
[[9]]$tags[[2]]
[[9]]$tags[[2]]$id
[1] 47624
[[9]]$tags[[2]]$name
[1] "R"
[[9]]$tags[[3]]
[[9]]$tags[[3]]$id
[1] 42046
[[9]]$tags[[3]]$name
[1] "r"
[[9]]$categories
[[9]]$categories[[1]]
[[9]]$categories[[1]]$id
[1] 77
[[9]]$categories[[1]]$name
[1] "Applied Computer Science"
[[9]]$files
[[9]]$files[[1]]
[[9]]$files[[1]]$download_url
[1] "http://files.figshare.com/98377/fs_create.R"
[[9]]$files[[1]]$size
[1] "2 KB"
[[9]]$files[[1]]$id
[1] 98377
[[9]]$files[[1]]$mime_type
[1] "text/plain"
[[9]]$files[[1]]$name
[1] "fs_create.R"
[[9]]$links
list()
[[10]]
[[10]]$article_id
[1] 138
[[10]]$title
[1] "Labrid adaptive peaks"
[[10]]$defined_type
[1] "figure"
[[10]]$status
[1] "Public"
[[10]]$version
[1] 2
[[10]]$published_date
[1] "13:45, Dec 30, 2011"
[[10]]$description
[1] "<p>Described in the notebook: http://openwetware.org/wiki/User:Carl_Boettiger/Notebook/Comparative_Phylogenetics/2010/03/12</p>"
[[10]]$description_nohtml
[1] "Described in the notebook: http://openwetware.org/wiki/User:Carl_Boettiger/Notebook/Comparative_Phylogenetics/2010/03/12"
[[10]]$total_size
[1] "29.71 KB"
[[10]]$authors
[[10]]$authors[[1]]
[[10]]$authors[[1]]$first_name
[1] "Carl"
[[10]]$authors[[1]]$last_name
[1] "Boettiger"
[[10]]$authors[[1]]$id
[1] 96387
[[10]]$authors[[1]]$full_name
[1] "Carl Boettiger"
[[10]]$tags
[[10]]$tags[[1]]
[[10]]$tags[[1]]$id
[1] 277
[[10]]$tags[[1]]$name
[1] "comparative methods"
[[10]]$tags[[2]]
[[10]]$tags[[2]]$id
[1] 276
[[10]]$tags[[2]]$name
[1] "phylogenetics"
[[10]]$tags[[3]]
[[10]]$tags[[3]]$id
[1] 275
[[10]]$tags[[3]]$name
[1] "fins"
[[10]]$tags[[4]]
[[10]]$tags[[4]]$id
[1] 274
[[10]]$tags[[4]]$name
[1] "labrids"
[[10]]$categories
[[10]]$categories[[1]]
[[10]]$categories[[1]]$id
[1] 24
[[10]]$categories[[1]]$name
[1] "Evolutionary biology"
[[10]]$categories[[2]]
[[10]]$categories[[2]]$id
[1] 39
[[10]]$categories[[2]]$name
[1] "Ecology"
[[10]]$files
[[10]]$files[[1]]
[[10]]$files[[1]]$download_url
[1] "http://files.figshare.com/137/Labrid_fins.png"
[[10]]$files[[1]]$size
[1] "30 KB"
[[10]]$files[[1]]$id
[1] 137
[[10]]$files[[1]]$mime_type
[1] "image/png"
[[10]]$files[[1]]$name
[1] "Labrid_fins.png"
[[10]]$links
list()
Note that we can easily grab the ids with the wrapper function fs_ids:
fs_ids(all_mine)
[1] 105136 105135 97653 97500 97279 97218 96919 96916 95839 138
We can delete unwanted files that are not public with fs_delete:
fs_delete(id)
Try the rfigshare package in your browser
Any scripts or data that you put into this service are public.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
