paper/paper.md
In AntonelliLab/outsider: Install and Run Programs, Outside of R, Inside of R
title: "outsider: Install and run programs, outside of R, inside of R"
tags:
- R
- Docker
- GitHub
- GitLab
- BitBucket
- Reproducibility
- Workflow
- Integration
authors:
- name: Dominic J. Bennett
orcid: 0000-0003-2722-1359
affiliation: "1, 2"
- name: Hannes Hettling
orcid: 0000-0003-4144-2238
affiliation: "3"
- name: Daniele Silvestro
orcid: 0000-0003-0100-0961
affiliation: "1, 2"
- name: Rutger Vos
orcid: 0000-0001-9254-7318
affiliation: "3"
- name: Alexandre Antonelli
orcid: 0000-0003-1842-9297
affiliation: "1, 2, 4"
affiliations:
- name: Gothenburg Global Biodiversity Centre, Box 461, SE-405 30 Gothenburg, Sweden
index: 1
- name: Department of Biological and Environmental Sciences, University of Gothenburg, Box 461, SE-405 30 Gothenburg, Sweden
index: 2
- name: Naturalis Biodiversity Center, P.O. Box 9517, 2300 RA Leiden, The Netherlands
index: 3
- name: Royal Botanic Gardens, Kew, TW9 3AE, Richmond, Surrey, UK
index: 4
date: 29 January 2020
bibliography: paper.bib
Statement of need
Enable integration of R and non-R code and programs to facilitate reproducible workflows.
Summary
In many areas of research, product development and software engineering, analytical pipelines – workflows connecting output from multiple software – are key for processing and running tests on data.
They can provide results in a consistent, modular and transparent manner.
Pipelines also make it easier to demonstrate the reproducibility of one’s research as well as enabling analyses that update as new data are added.
Not all analyses, however, can necessarily be run or coded in one’s favoured programming language as different parts of an analysis may require external software or packages.
Integrating a variety of programs and software can lead to issues of portability (additional software may not run across all operating systems) and versioning errors (differing arguments across additional software versions).
For the ideal pipeline, it should be possible to install and run any command-line software, within the main programming language of the pipeline, without concern for software versions or operating system.
R [@cran] is one of the most popular computer languages amongst researchers, and many packages exist for calling programs and code from non-R sources (e.g. sys [@sys] for shell commands, reticulate [@reticulate] for python and rJava [@rJava] for Java).
To our knowledge, however, no R package exists with the ability to launch external programs originating from any UNIX command-line source.
The outsider packages work through docker [@docker] – a service that, through OS-level virtualization, enables deployment of isolated software "containers" – and a code-sharing service, e.g. GitHub [@github], to allow a user to install and run, in theory, any external, command-line program or package, on any of the major operating systems (Windows, Linux, OSX).
How it works
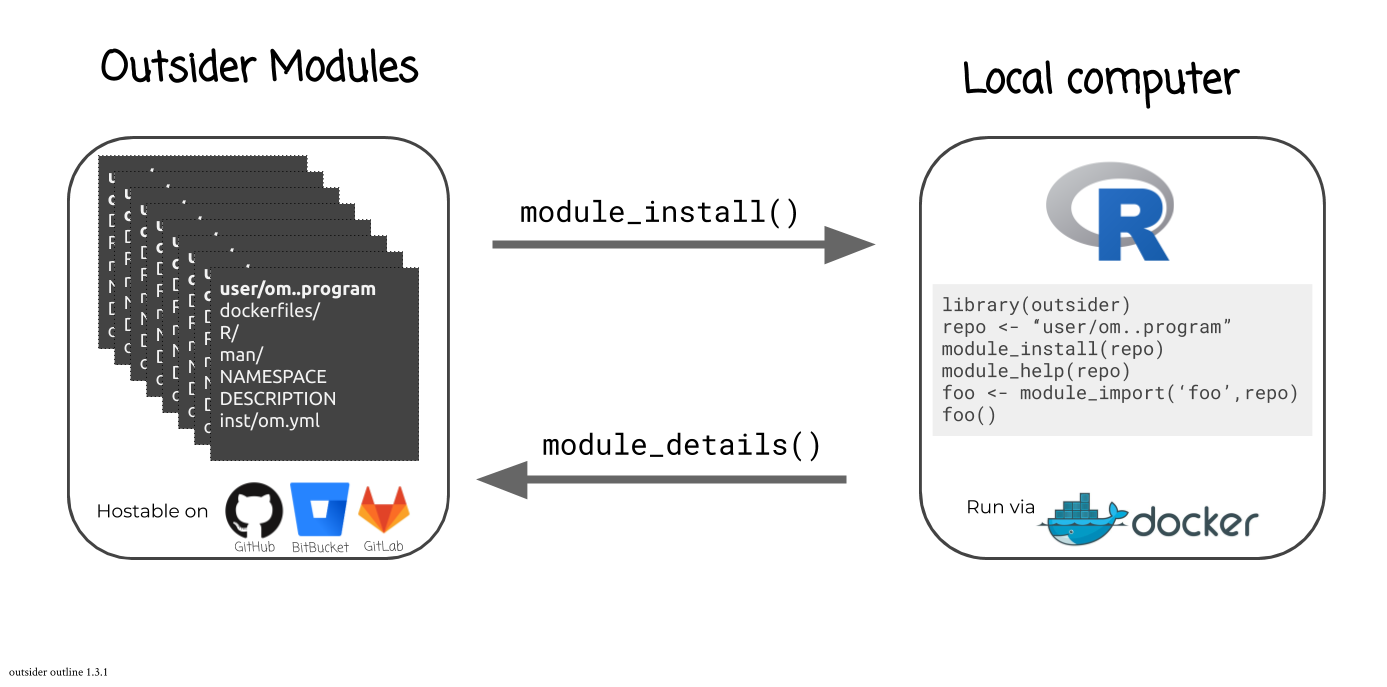
outsider packages provide an interface to install and run outsider modules.
These modules are hostable on GitHub [@github], GitLab [@gitlab] and/or BitBucket [@bitbucket] and consist of two parts: a (barebones) R package and a Dockerfile.
The Dockerfile details the installation process for an external program contained within a Docker image, while the R package comprises functions and documentation for interacting with the external program via a Docker container.
For many programs, Dockerfiles are readily available online and require minor changes to adapt for outsider.
By default, a module’s R code simply passes command-line arguments through Docker.
After installation, a module’s functions can then be imported and launched using outsider functions.
Upon running a module’s code, outsider code will first launch a Docker container of the image as described by the module’s Dockerfile.
outsider then facilitates the communication between the module’s R code and the Docker container that hosts the external program (developers of modules have the choice of determining default behaviours for handling generated files).
outsider modules thus wrap external command-line programs into R functions in a convenient manner.
outsider functions allow users to look up available modules and determine build statuses (i.e. whether the package is passing its online tests) before installing.
At time of writing, outsider modules for some of the most popular bioinformatics tools have been developed: BLAST [@blast], MAFFT [@mafft], *BEAST [@beast], RAxML [@raxml], bamm [@bamm], PyRate [@pyrate].
(See the outsider website for an up-to-date and complete list).
All that is required to run these modules is R and Docker.
Docker Desktop [@docker_desktop] can be installed for all operating systems but for older versions of OSX and Windows the legacy "Docker Toolbox" [@docker_toolbox] may instead need to be installed.
(Note, users may need to create an account with Docker-Hub to install Docker.)
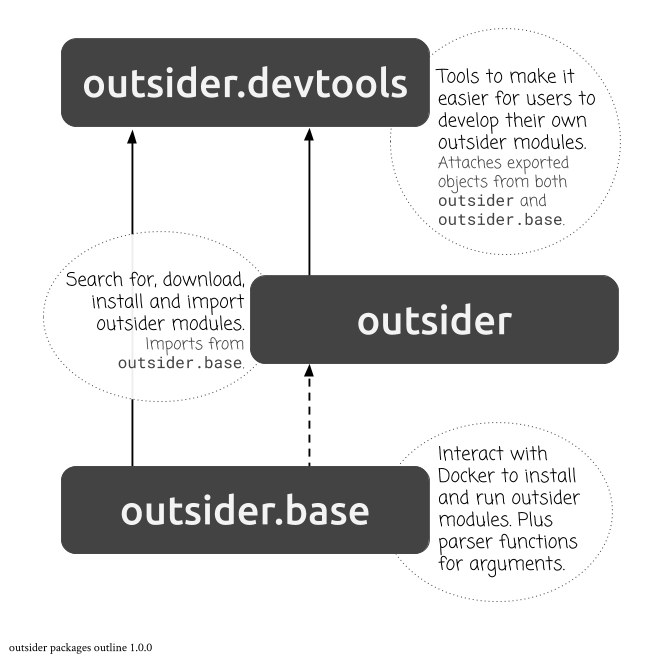
Code structure
The code-base that allows for the installation, execution and development of outsider modules is held across three different R packages. For end-users of modules, however, only the outsider module is required. For those who wish to develop their own modules, the outsider.devtools package provides helper functions for doing so. In addition, there is a test suites repository that hosts mock analysis pipelines that initiate several modules in sequence to test the interaction of all the packages.
outsider: The main package for installing, importing and running outsider modules [@outsider_z].outsider.base: The package for low-level interaction between module R code and Docker containers (not user-facing) [@outsiderbase_z].outsider.devtools: The development tools package for facilitating the creation of new modules [@outsiderdev_z].- "outsider-testuites": A repository hosting a series of "test" pipelines for ensuring modules can be successfully strung together to form R/non-R workflows [@outsiderts_z].
Examples
Saying hello from Ubuntu
By hosting a Docker container, outsider can run any UNIX-based, external command-line program.
To demonstrate this process we can say "hello world" via a container hosting the Ubuntu operating system.
In this short example, we will install a small outsider module – om..hello.world – that installs a local copy of the latest version of Ubuntu and contains a function for saying hello using the command echo.
library(outsider)
# outsider modules are hosted on GitHub
# this repo is a demonstration of an outsider module
# it contains a function for printing 'Hello World!'
repo <- 'dombennett/om..hello.world'
module_install(repo = repo)
# look up the help files for the module
module_help(repo = repo)
# import the 'hello_world' function
hello_world <- module_import(fname = 'hello_world', repo = repo)
# run the imported function
hello_world()
#> Hello world!
#> ------------
#> DISTRIB_ID=Ubuntu
#> DISTRIB_RELEASE=18.04
#> DISTRIB_CODENAME=bionic
#> DISTRIB_DESCRIPTION="Ubuntu 18.04.1 LTS"
A basic bioinformatic pipeline
To better demonstrate the power of the outsider package, we will run a simple bioinformatic pipeline that downloads a file of biological sequence data (borrowed from [@genomeworkshop]) and aligns the separate strands of DNA using the multiple sequence alignment program MAFFT [@mafft].
Note that we can pass arguments to an outsider module, such as mafft in the example below, using separate R arguments for each command-line argument.
library(outsider)
repo <- 'dombennett/om..mafft'
module_install(repo = repo)
mafft <- module_import(fname = 'mafft', repo = repo)
# some example file
download.file('https://molb7621.github.io/workshop/_downloads/sample.fa',
'sample.fa')
# run maft with --auto and write results to alignment.fa
mafft(arglist = c('--auto', 'sample.fa', '>', 'alignment.fa'))
# view alignment
cat(readLines('alignment.fa'), sep = '\n')
#> >derice
#> -actgactagctagctaactg
#> >sanka
#> -gcatcgtagctagctacgat
#> >junior
#> catcgatcgtacgtacg-tag
#> >yul
#> -atcgatcgatcgtacgatcg
For more detailed and up-to-date examples and tutorials, see the outsider GitHub page [@outsider_gh].
Availability
outsider (and its sister packages) are open-source software made available under the MIT licence allowing reuse of the software with limited constraints.
It is aimed that all packages will be made available through CRAN [@cran], e.g. install.package("outsider").
Currently, all are available from GitHub source code repositories using the remotes package, e.g. remotes::install_github("ropensci/outsider")
Funding
This package has been developed as part of the supersmartR project [@supersmartR] which has received funding through A.A. (from the Swedish Research Council [B0569601], the Swedish Foundation for Strategic Research and a Wallenberg Academy Fellowship) and through D.S. (from the Swedish Research Council [2015-04748]).
Acknowledgements
The authors wish to thank Daniel Nüst and Bob Rudis for taking the time to review the package and providing valuable ideas and suggestions.
Additionally, we would like to thank Anna Krystalli for overseeing the review process and providing useful advice in turn.
Finally, we would like to thank all the people behind ROpenSci and JOSS for making this project and its review possible.
References
AntonelliLab/outsider documentation built on June 17, 2022, 3:27 p.m.
title: "outsider: Install and run programs, outside of R, inside of R" tags: - R - Docker - GitHub - GitLab - BitBucket - Reproducibility - Workflow - Integration authors: - name: Dominic J. Bennett orcid: 0000-0003-2722-1359 affiliation: "1, 2" - name: Hannes Hettling orcid: 0000-0003-4144-2238 affiliation: "3" - name: Daniele Silvestro orcid: 0000-0003-0100-0961 affiliation: "1, 2" - name: Rutger Vos orcid: 0000-0001-9254-7318 affiliation: "3" - name: Alexandre Antonelli orcid: 0000-0003-1842-9297 affiliation: "1, 2, 4" affiliations: - name: Gothenburg Global Biodiversity Centre, Box 461, SE-405 30 Gothenburg, Sweden index: 1 - name: Department of Biological and Environmental Sciences, University of Gothenburg, Box 461, SE-405 30 Gothenburg, Sweden index: 2 - name: Naturalis Biodiversity Center, P.O. Box 9517, 2300 RA Leiden, The Netherlands index: 3 - name: Royal Botanic Gardens, Kew, TW9 3AE, Richmond, Surrey, UK index: 4 date: 29 January 2020 bibliography: paper.bib
Statement of need
Enable integration of R and non-R code and programs to facilitate reproducible workflows.
Summary
In many areas of research, product development and software engineering, analytical pipelines – workflows connecting output from multiple software – are key for processing and running tests on data.
They can provide results in a consistent, modular and transparent manner.
Pipelines also make it easier to demonstrate the reproducibility of one’s research as well as enabling analyses that update as new data are added.
Not all analyses, however, can necessarily be run or coded in one’s favoured programming language as different parts of an analysis may require external software or packages.
Integrating a variety of programs and software can lead to issues of portability (additional software may not run across all operating systems) and versioning errors (differing arguments across additional software versions).
For the ideal pipeline, it should be possible to install and run any command-line software, within the main programming language of the pipeline, without concern for software versions or operating system.
R [@cran] is one of the most popular computer languages amongst researchers, and many packages exist for calling programs and code from non-R sources (e.g. sys [@sys] for shell commands, reticulate [@reticulate] for python and rJava [@rJava] for Java).
To our knowledge, however, no R package exists with the ability to launch external programs originating from any UNIX command-line source.
The outsider packages work through docker [@docker] – a service that, through OS-level virtualization, enables deployment of isolated software "containers" – and a code-sharing service, e.g. GitHub [@github], to allow a user to install and run, in theory, any external, command-line program or package, on any of the major operating systems (Windows, Linux, OSX).
How it works
outsider packages provide an interface to install and run outsider modules.
These modules are hostable on GitHub [@github], GitLab [@gitlab] and/or BitBucket [@bitbucket] and consist of two parts: a (barebones) R package and a Dockerfile.
The Dockerfile details the installation process for an external program contained within a Docker image, while the R package comprises functions and documentation for interacting with the external program via a Docker container.
For many programs, Dockerfiles are readily available online and require minor changes to adapt for outsider.
By default, a module’s R code simply passes command-line arguments through Docker.
After installation, a module’s functions can then be imported and launched using outsider functions.
Upon running a module’s code, outsider code will first launch a Docker container of the image as described by the module’s Dockerfile.
outsider then facilitates the communication between the module’s R code and the Docker container that hosts the external program (developers of modules have the choice of determining default behaviours for handling generated files).
outsider modules thus wrap external command-line programs into R functions in a convenient manner.
outsider functions allow users to look up available modules and determine build statuses (i.e. whether the package is passing its online tests) before installing.
At time of writing, outsider modules for some of the most popular bioinformatics tools have been developed: BLAST [@blast], MAFFT [@mafft], *BEAST [@beast], RAxML [@raxml], bamm [@bamm], PyRate [@pyrate].
(See the outsider website for an up-to-date and complete list).
All that is required to run these modules is R and Docker.
Docker Desktop [@docker_desktop] can be installed for all operating systems but for older versions of OSX and Windows the legacy "Docker Toolbox" [@docker_toolbox] may instead need to be installed.
(Note, users may need to create an account with Docker-Hub to install Docker.)

Code structure
The code-base that allows for the installation, execution and development of outsider modules is held across three different R packages. For end-users of modules, however, only the outsider module is required. For those who wish to develop their own modules, the outsider.devtools package provides helper functions for doing so. In addition, there is a test suites repository that hosts mock analysis pipelines that initiate several modules in sequence to test the interaction of all the packages.
outsider: The main package for installing, importing and running outsider modules [@outsider_z].outsider.base: The package for low-level interaction between module R code and Docker containers (not user-facing) [@outsiderbase_z].outsider.devtools: The development tools package for facilitating the creation of new modules [@outsiderdev_z].- "outsider-testuites": A repository hosting a series of "test" pipelines for ensuring modules can be successfully strung together to form R/non-R workflows [@outsiderts_z].

Examples
Saying hello from Ubuntu
By hosting a Docker container, outsider can run any UNIX-based, external command-line program.
To demonstrate this process we can say "hello world" via a container hosting the Ubuntu operating system.
In this short example, we will install a small outsider module – om..hello.world – that installs a local copy of the latest version of Ubuntu and contains a function for saying hello using the command echo.
library(outsider)
# outsider modules are hosted on GitHub
# this repo is a demonstration of an outsider module
# it contains a function for printing 'Hello World!'
repo <- 'dombennett/om..hello.world'
module_install(repo = repo)
# look up the help files for the module
module_help(repo = repo)
# import the 'hello_world' function
hello_world <- module_import(fname = 'hello_world', repo = repo)
# run the imported function
hello_world()
#> Hello world!
#> ------------
#> DISTRIB_ID=Ubuntu
#> DISTRIB_RELEASE=18.04
#> DISTRIB_CODENAME=bionic
#> DISTRIB_DESCRIPTION="Ubuntu 18.04.1 LTS"
A basic bioinformatic pipeline
To better demonstrate the power of the outsider package, we will run a simple bioinformatic pipeline that downloads a file of biological sequence data (borrowed from [@genomeworkshop]) and aligns the separate strands of DNA using the multiple sequence alignment program MAFFT [@mafft].
Note that we can pass arguments to an outsider module, such as mafft in the example below, using separate R arguments for each command-line argument.
library(outsider)
repo <- 'dombennett/om..mafft'
module_install(repo = repo)
mafft <- module_import(fname = 'mafft', repo = repo)
# some example file
download.file('https://molb7621.github.io/workshop/_downloads/sample.fa',
'sample.fa')
# run maft with --auto and write results to alignment.fa
mafft(arglist = c('--auto', 'sample.fa', '>', 'alignment.fa'))
# view alignment
cat(readLines('alignment.fa'), sep = '\n')
#> >derice
#> -actgactagctagctaactg
#> >sanka
#> -gcatcgtagctagctacgat
#> >junior
#> catcgatcgtacgtacg-tag
#> >yul
#> -atcgatcgatcgtacgatcg
For more detailed and up-to-date examples and tutorials, see the outsider GitHub page [@outsider_gh].
Availability
outsider (and its sister packages) are open-source software made available under the MIT licence allowing reuse of the software with limited constraints.
It is aimed that all packages will be made available through CRAN [@cran], e.g. install.package("outsider").
Currently, all are available from GitHub source code repositories using the remotes package, e.g. remotes::install_github("ropensci/outsider")
Funding
This package has been developed as part of the supersmartR project [@supersmartR] which has received funding through A.A. (from the Swedish Research Council [B0569601], the Swedish Foundation for Strategic Research and a Wallenberg Academy Fellowship) and through D.S. (from the Swedish Research Council [2015-04748]).
Acknowledgements
The authors wish to thank Daniel Nüst and Bob Rudis for taking the time to review the package and providing valuable ideas and suggestions. Additionally, we would like to thank Anna Krystalli for overseeing the review process and providing useful advice in turn. Finally, we would like to thank all the people behind ROpenSci and JOSS for making this project and its review possible.
References
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
