In AntonelliLab/outsider: Install and Run Programs, Outside of R, Inside of R
Many data science, statistical and academic projects require running models on
external software and then performing subsequent analyses of the results in R
e.g. for project specific testing and visualisation. outsider aims to make
this process simpler by first enabling non-R software to be run from within R
and, second, by making it easier to install external program.
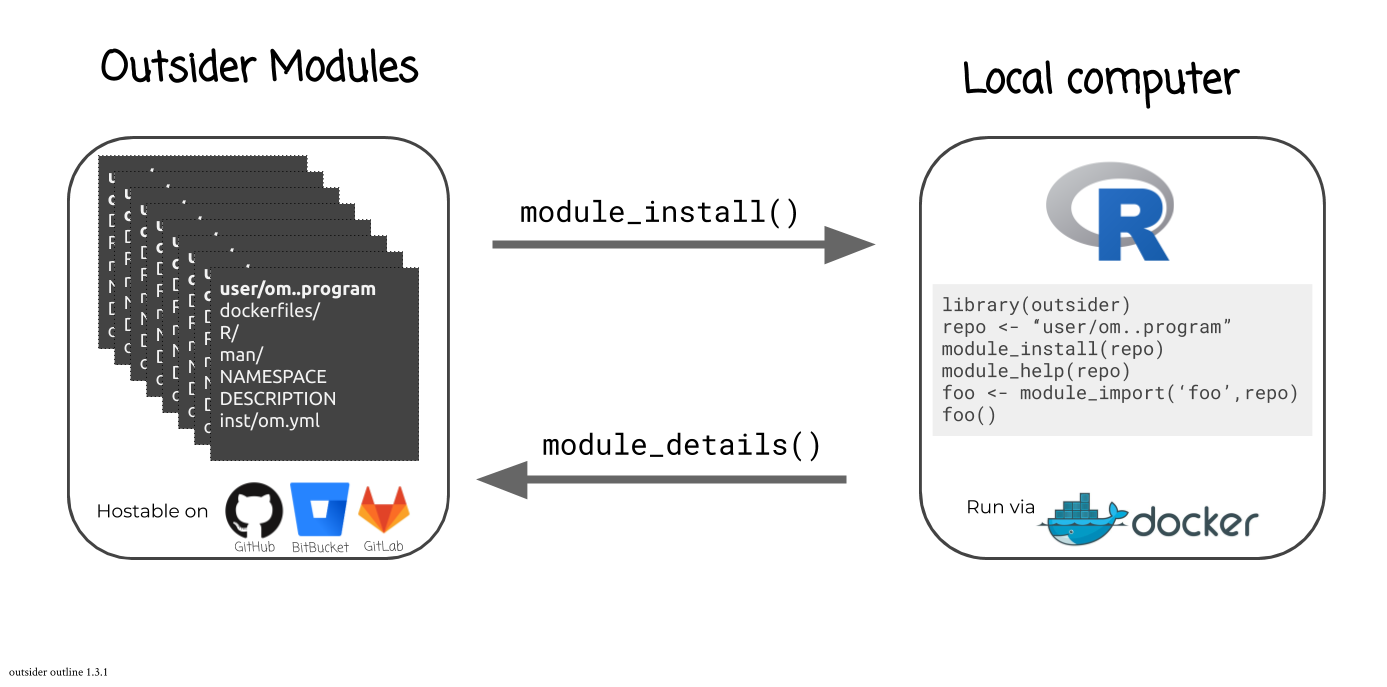
How it works
The outsider package acts as an interface between the R environment and
external programs that are hosted on virtual machines. A virtual machine is
hosted on a user's computer but acts like an external computer with its own
operating system. These virtual machines are run through the program Docker. So long as a computer is running Docker,
then any of these virtual machines can be downloaded and run without any
installation process. Docker runs on multiple operating systems including
Windows, OSX and Linux.
For every external program provided through outsider, a virtual machine, or
"Docker image", needs to be described and specific R code -- for launching and
interacting with the program -- is required. This Docker image and R code are
provided through outsider modules that are hosted on
GitHub.
Users can install any of the available outsider modules. With two commands,
a user can install and import an external program for calling within R:
module_install() and module_import().
Installing outsider
Before you can make the most of outsider you will need to install and start
running Docker. Follow the installation instructions for your specific operating
system, "Install Docker".
For some operating systems, "Docker Desktop" is not available. If that is the
case, try "Docker Toolbox". This is a
legacy Docker for older operating systems. It has similar functionality but
requires a virtual machine and has greater computational overhead.
With Docker installed, you then can install outsider via GitHub.
library(remotes)
install_remotes('ropensci/outsider')
Non-R dependencies
- For Windows users you may need to install
Rtools.
- For Mac users you may need to install and setup
"Xcode command-line tools", by running the command
xcode-select --install in
the terminal.
Finding and installing modules
To see what modules are available you can see the "available modules page".
Alternatively, for the latest available information you can search for modules
using the module_details() function.
library(outsider)
# repo = NULL will search for ALL available modules
# (this may take a long time, depends on internet connection and remote server)
print(module_details(repo = 'dombennett/om..mafft'))
## # A tibble: 1 x 7
## repo program details versions updated_at watchers_count url
## <chr> <chr> <chr> <chr> <dttm> <int> <chr>
## 1 dombennett/… mafft Multiple alignment program for… latest 2020-01-16 10:36:00 0 https://github.co…
To install a module, all that is required is to provide the repo name to the
function module_install().
library(outsider)
module_install(repo = 'dombennett/om..mafft', force = TRUE)
What is repo? The repo is the unique name for a GitHub repository that
hosts an outsider module. It consists of two parts: a GitHub username and a
project name. Given its uniqueness, all modules are referred to by their repo.
To confirm the module is installed on a computer, it might be useful to use
module_installed(). This function returns a table of all installed modules.
library(outsider)
print(module_installed())
## # A tibble: 3 x 7
## package image tag program url image_created image_id
## <fct> <chr> <chr> <chr> <chr> <chr> <chr>
## 1 om..hello.world dombennett/om_hello_… latest hello world https://github.com/DomBennett/o… 8 months ago acdff0a24…
## 2 om..mafft dombennett/om_mafft latest mafft https://github.com/DomBennett/o… 13 months ago 97170a5f7…
## 3 om..partitionf… dombennett/om_partit… <NA> PartitionFi… https://github.com/DomBennett/o… <NA> <NA>
Importing
All modules contain functions for interacting with the external program that
they host. To see these functions we can use module_help() to look up the help
documents.
library(outsider)
# the whole module
module_help(repo = 'dombennett/om..mafft')
# specific function of a module (if known)
module_help(repo = 'dombennett/om..mafft', fname = 'mafft')
Once a function name is known of a particular module, the function can be
imported with module_import().
library(outsider)
mafft <- module_import(fname = 'mafft', repo = 'dombennett/om..mafft')
print(is(mafft))
## [1] "function" "OptionalFunction" "PossibleMethod"
What is mafft? "mafft" is a multiple alignment tool for for biological
sequences. Note, a user can use any name they wish for the function when it
is imported. For example, mafftymcmafftface <- module_import( ... would work
equally well.
Commands
The imported functions from modules act like portals to the external programs
the modules host. To run a command, a user needs to use the function name and
give arguments corresponding to the arguments of the external program. For
example, on command-line to list the help information for mafft, we would
write mafft --help. With outsider we can do run mafft('--help').
For a more complicated example, we could launch a small analysis with mafft
like so.
library(outsider)
mafft <- module_import(fname = 'mafft', repo = 'dombennett/om..mafft')
mafft(arglist = c('--auto', 'input_sequences.fasta', '>',
'output_alignment.fasta'))
Why the spaces between arguments? All the arguments of the external,
command-line program must be provided as separated characters. This helps
outsider parse the elements.
How does that last line work?
mafft(arglist = c('--auto', 'input_sequences.fasta', '>',
'output_alignment.fasta'))
describes in R how to call the MAFFT program via command-line/terminal. It is
equivalent to mafft --auto input_sequences.fasta > output_alignment.fasta if
we were to call the program via command-line/terminal. How do we know how to
structure the program arguments? In the case of MAFFT we can look-up the
arguments on their website, mafft.cbrc.jp. But often for
command-line programs we can call for help with -h or --help. For MAFFT at
the command-line, we could run mafft --help or with outsider we can run:
mafft(arglist = '--help')
##
------------------------------------------------------------------------------
## MAFFT v7.407 (2018/Jul/23)
##
-
https://mafft.cbrc.jp/alignment/software/
## MBE 30:772-780 (2013), NAR 30:3059-3066 (2002)
## ------------------------------------------------------------------------------
## High speed:
## % mafft in > out
## % mafft --retree 1 in > out (fast)
##
## High accuracy (for <~200 sequences x <~2,000 aa/nt):
## % mafft --maxiterate 1000 --localpair in > out (% linsi in > out is also ok)
## % mafft --maxiterate 1000 --genafpair in > out (% einsi in > out)
## % mafft --maxiterate 1000 --globalpair in > out (% ginsi in > out)
##
## If unsure which option to use:
## % mafft --auto in > out
##
## --op # : Gap opening penalty, default: 1.53
## --ep # : Offset (works like gap extension penalty), default: 0.0
## --maxiterate # : Maximum number of iterative refinement, default: 0
## --clustalout : Output: clustal format, default: fasta
## --reorder : Outorder: aligned, default: input order
## --quiet : Do not report progress
## --thread # : Number of threads (if unsure, --thread -1)
##
The help page returned tells us how to structure the arguments:
[options] [input_file] > [output_file]
Where the options (e.g. alignment method, number of threads) are always
indicated first with -- and the input and output files are indicated second
with the >.
Uninstalling
Clean up your computer by removing unwanted modules with module_uninstall().
library(outsider)
module_uninstall(repo = 'dombennett/om..mafft')
Building your own module
Unfortunately, outsider's utility is limited by the number of available
modules. Fortunately, it is very easy to create and upload your own module.
The package comes with a range of helper functions for minimising the amount of
coding for a module developer. If you know how to install an external program on
your own computer which you would like would like to run it through outsider
and you have some experience with GitHub, then explore the
"outsider.devtools"
package.
AntonelliLab/outsider documentation built on June 17, 2022, 3:27 p.m.
Many data science, statistical and academic projects require running models on
external software and then performing subsequent analyses of the results in R
e.g. for project specific testing and visualisation. outsider aims to make
this process simpler by first enabling non-R software to be run from within R
and, second, by making it easier to install external program.
How it works
The outsider package acts as an interface between the R environment and
external programs that are hosted on virtual machines. A virtual machine is
hosted on a user's computer but acts like an external computer with its own
operating system. These virtual machines are run through the program Docker. So long as a computer is running Docker,
then any of these virtual machines can be downloaded and run without any
installation process. Docker runs on multiple operating systems including
Windows, OSX and Linux.
For every external program provided through outsider, a virtual machine, or
"Docker image", needs to be described and specific R code -- for launching and
interacting with the program -- is required. This Docker image and R code are
provided through outsider modules that are hosted on
GitHub.
Users can install any of the available outsider modules. With two commands,
a user can install and import an external program for calling within R:
module_install() and module_import().

Installing outsider
Before you can make the most of outsider you will need to install and start
running Docker. Follow the installation instructions for your specific operating
system, "Install Docker".
For some operating systems, "Docker Desktop" is not available. If that is the case, try "Docker Toolbox". This is a legacy Docker for older operating systems. It has similar functionality but requires a virtual machine and has greater computational overhead.
With Docker installed, you then can install outsider via GitHub.
library(remotes) install_remotes('ropensci/outsider')
Non-R dependencies
- For Windows users you may need to install Rtools.
- For Mac users you may need to install and setup
"Xcode command-line tools", by running the command
xcode-select --installin the terminal.
Finding and installing modules
To see what modules are available you can see the "available modules page".
Alternatively, for the latest available information you can search for modules
using the module_details() function.
library(outsider) # repo = NULL will search for ALL available modules # (this may take a long time, depends on internet connection and remote server) print(module_details(repo = 'dombennett/om..mafft'))
## # A tibble: 1 x 7 ## repo program details versions updated_at watchers_count url ## <chr> <chr> <chr> <chr> <dttm> <int> <chr> ## 1 dombennett/… mafft Multiple alignment program for… latest 2020-01-16 10:36:00 0 https://github.co…
To install a module, all that is required is to provide the repo name to the
function module_install().
library(outsider) module_install(repo = 'dombennett/om..mafft', force = TRUE)
What is
repo? Therepois the unique name for a GitHub repository that hosts anoutsidermodule. It consists of two parts: a GitHub username and a project name. Given its uniqueness, all modules are referred to by theirrepo.
To confirm the module is installed on a computer, it might be useful to use
module_installed(). This function returns a table of all installed modules.
library(outsider) print(module_installed())
## # A tibble: 3 x 7 ## package image tag program url image_created image_id ## <fct> <chr> <chr> <chr> <chr> <chr> <chr> ## 1 om..hello.world dombennett/om_hello_… latest hello world https://github.com/DomBennett/o… 8 months ago acdff0a24… ## 2 om..mafft dombennett/om_mafft latest mafft https://github.com/DomBennett/o… 13 months ago 97170a5f7… ## 3 om..partitionf… dombennett/om_partit… <NA> PartitionFi… https://github.com/DomBennett/o… <NA> <NA>
Importing
All modules contain functions for interacting with the external program that
they host. To see these functions we can use module_help() to look up the help
documents.
library(outsider) # the whole module module_help(repo = 'dombennett/om..mafft') # specific function of a module (if known) module_help(repo = 'dombennett/om..mafft', fname = 'mafft')
Once a function name is known of a particular module, the function can be
imported with module_import().
library(outsider) mafft <- module_import(fname = 'mafft', repo = 'dombennett/om..mafft') print(is(mafft))
## [1] "function" "OptionalFunction" "PossibleMethod"
What is
mafft? "mafft" is a multiple alignment tool for for biological sequences. Note, a user can use any name they wish for the function when it is imported. For example,mafftymcmafftface <- module_import( ...would work equally well.
Commands
The imported functions from modules act like portals to the external programs
the modules host. To run a command, a user needs to use the function name and
give arguments corresponding to the arguments of the external program. For
example, on command-line to list the help information for mafft, we would
write mafft --help. With outsider we can do run mafft('--help').
For a more complicated example, we could launch a small analysis with mafft
like so.
library(outsider) mafft <- module_import(fname = 'mafft', repo = 'dombennett/om..mafft') mafft(arglist = c('--auto', 'input_sequences.fasta', '>', 'output_alignment.fasta'))
Why the spaces between arguments? All the arguments of the external, command-line program must be provided as separated characters. This helps
outsiderparse the elements.
How does that last line work?
mafft(arglist = c('--auto', 'input_sequences.fasta', '>',
'output_alignment.fasta'))
describes in R how to call the MAFFT program via command-line/terminal. It is
equivalent to mafft --auto input_sequences.fasta > output_alignment.fasta if
we were to call the program via command-line/terminal. How do we know how to
structure the program arguments? In the case of MAFFT we can look-up the
arguments on their website, mafft.cbrc.jp. But often for
command-line programs we can call for help with -h or --help. For MAFFT at
the command-line, we could run mafft --help or with outsider we can run:
mafft(arglist = '--help')
## ------------------------------------------------------------------------------ ## MAFFT v7.407 (2018/Jul/23) ## - https://mafft.cbrc.jp/alignment/software/ ## MBE 30:772-780 (2013), NAR 30:3059-3066 (2002) ## ------------------------------------------------------------------------------ ## High speed: ## % mafft in > out ## % mafft --retree 1 in > out (fast) ## ## High accuracy (for <~200 sequences x <~2,000 aa/nt): ## % mafft --maxiterate 1000 --localpair in > out (% linsi in > out is also ok) ## % mafft --maxiterate 1000 --genafpair in > out (% einsi in > out) ## % mafft --maxiterate 1000 --globalpair in > out (% ginsi in > out) ## ## If unsure which option to use: ## % mafft --auto in > out ## ## --op # : Gap opening penalty, default: 1.53 ## --ep # : Offset (works like gap extension penalty), default: 0.0 ## --maxiterate # : Maximum number of iterative refinement, default: 0 ## --clustalout : Output: clustal format, default: fasta ## --reorder : Outorder: aligned, default: input order ## --quiet : Do not report progress ## --thread # : Number of threads (if unsure, --thread -1) ##
The help page returned tells us how to structure the arguments:
[options] [input_file] > [output_file]
Where the options (e.g. alignment method, number of threads) are always
indicated first with -- and the input and output files are indicated second
with the >.
Uninstalling
Clean up your computer by removing unwanted modules with module_uninstall().
library(outsider) module_uninstall(repo = 'dombennett/om..mafft')
Building your own module
Unfortunately, outsider's utility is limited by the number of available
modules. Fortunately, it is very easy to create and upload your own module.
The package comes with a range of helper functions for minimising the amount of
coding for a module developer. If you know how to install an external program on
your own computer which you would like would like to run it through outsider
and you have some experience with GitHub, then explore the
"outsider.devtools"
package.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
