In DHLab-CGU/emr: An analysis tool for diagnosis and procedure records
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>"
)
Getting started with dxpr package

I. Introduction
The proposed open-source dxpr package is a software tool aimed at expediting an integrated analysis of electronic health records (EHRs). The dxpr package provides mechanisms to integrate, analyze, and visualize clinical data, including diagnosis and procedure records.
Feature
- Data integration Transform codes into uniform format and group code into several categories.
- Data Wrangling Generate statistical information about dataset and transform data into wide format, which fits better to other analytical and plotting packages.
- Visualization Provide overviews for the result of diagnoses standardization and the grouped categories of diagnosis codes.
Getting started
- Diagnostic part
English: https://dhlab-tseng.github.io/dxpr/articles/Eng_Diagnosis.html
Chinese: https://dhlab-tseng.github.io/dxpr/articles/Chi_Diagnosis.html
- Procedure part
English: https://dhlab-tseng.github.io/dxpr/articles/Eng_Procedure.html
Chinese: https://dhlab-tseng.github.io/dxpr/articles/Chi_Procedure.html
Development version
# install.packages("remotes")
remotes::install_github("DHLab-TSENG/dxpr")
knitr::opts_chunk$set(collapse = T, comment = "#>")
options(tibble.print_min = 4L, tibble.print_max = 4L)
remotes::install_github("DHLab-TSENG/dxpr")
Overview
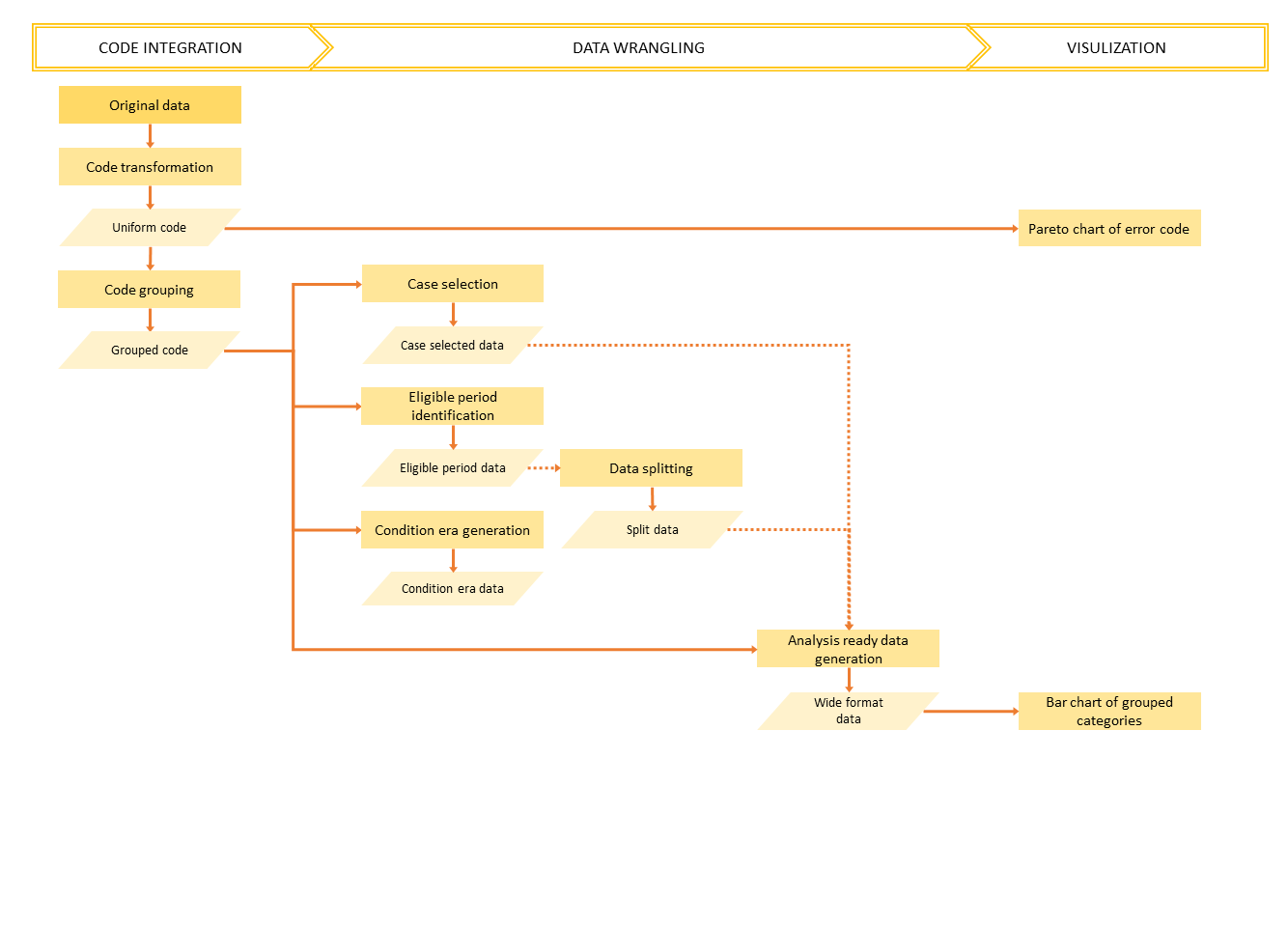
Usage
library(dxpr)
head(sampleDxFile)
# I. Data integration
# 1. Data standardization
short <- icdDxDecimalToShort(dxDataFile = sampleDxFile,
icdColName = ICD,
dateColName = Date,
icd10usingDate = "2015/10/01")
head(short$ICD)
tail(short$Error)
# 2. Data grouping
ELIX <- icdDxToComorbid(dxDataFile = sampleDxFile,
idColName = ID,
icdColName = ICD,
dateColName = Date,
icd10usingDate = "2015/10/01",
comorbidMethod = elix)
head(ELIX$groupedDT)
head(ELIX$summarised_groupedDT)
# II. Data wrangling
groupedDataWide <- groupedDataLongToWide(dxDataFile = ELIX$groupedDT,
idColName = ID,
categoryColName = Comorbidity,
dateColName = Date)
head(groupedDataWide[,1:4])
# IV. Visualization
plot_errorICD <- plotICDError(short$Error)
plot_groupedData <- plotDiagCat(groupedDataWide, ID)
plot_errorICD
plot_groupedData
Getting help
See the GitHub issues page (https://github.com/DHLab-TSENG/dxpr/issues) to see open issues and feature requests.
DHLab-CGU/emr documentation built on Sept. 2, 2023, 9:16 p.m.
knitr::opts_chunk$set( collapse = TRUE, comment = "#>" )
Getting started with dxpr package

I. Introduction
The proposed open-source dxpr package is a software tool aimed at expediting an integrated analysis of electronic health records (EHRs). The dxpr package provides mechanisms to integrate, analyze, and visualize clinical data, including diagnosis and procedure records.
Feature
- Data integration Transform codes into uniform format and group code into several categories.
- Data Wrangling Generate statistical information about dataset and transform data into wide format, which fits better to other analytical and plotting packages.
- Visualization Provide overviews for the result of diagnoses standardization and the grouped categories of diagnosis codes.
Getting started
- Diagnostic part
English: https://dhlab-tseng.github.io/dxpr/articles/Eng_Diagnosis.html
Chinese: https://dhlab-tseng.github.io/dxpr/articles/Chi_Diagnosis.html - Procedure part
English: https://dhlab-tseng.github.io/dxpr/articles/Eng_Procedure.html
Chinese: https://dhlab-tseng.github.io/dxpr/articles/Chi_Procedure.html
Development version
# install.packages("remotes") remotes::install_github("DHLab-TSENG/dxpr")
knitr::opts_chunk$set(collapse = T, comment = "#>") options(tibble.print_min = 4L, tibble.print_max = 4L) remotes::install_github("DHLab-TSENG/dxpr")
Overview

Usage
library(dxpr) head(sampleDxFile) # I. Data integration # 1. Data standardization short <- icdDxDecimalToShort(dxDataFile = sampleDxFile, icdColName = ICD, dateColName = Date, icd10usingDate = "2015/10/01") head(short$ICD) tail(short$Error) # 2. Data grouping ELIX <- icdDxToComorbid(dxDataFile = sampleDxFile, idColName = ID, icdColName = ICD, dateColName = Date, icd10usingDate = "2015/10/01", comorbidMethod = elix) head(ELIX$groupedDT) head(ELIX$summarised_groupedDT) # II. Data wrangling groupedDataWide <- groupedDataLongToWide(dxDataFile = ELIX$groupedDT, idColName = ID, categoryColName = Comorbidity, dateColName = Date) head(groupedDataWide[,1:4]) # IV. Visualization plot_errorICD <- plotICDError(short$Error) plot_groupedData <- plotDiagCat(groupedDataWide, ID) plot_errorICD plot_groupedData
Getting help
See the GitHub issues page (https://github.com/DHLab-TSENG/dxpr/issues) to see open issues and feature requests.
Embedding an R snippet on your website
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
