inst/examples/pomp-example.md
In cboettig/nonparametric-bayes: Nonparametric Bayes inference for ecological models
library(pomp)
## Loading required package: mvtnorm
## Loading required package: subplex
## Loading required package: deSolve
opts_knit$set(upload.fun = socialR::flickr.url)
options(flickr_tags = "nonparametric-bayes")
process.sim <- function(x, t, params, delta.t, ...) {
# r = 0.75, K = 10, Q = 3, H = 1, a = 1.3
r <- params["r"]
K <- params["K"]
Q <- params["Q"]
H <- params["H"]
a <- params["a"]
sigma <- params["sigma"]
X <- x["X"]
epsilon <- exp(rlnorm(n = 1, mean = 0, sd = sigma))
c(X = unname(X * exp(r * delta.t * (1 - X/K) - a * X^(Q - 1)/(X^Q + H^Q)) *
epsilon))
}
measure.sim <- function(x, t, params, ...) {
tau <- params["tau"]
X <- x["X"]
y <- c(Y = unname(rlnorm(n = 1, meanlog = log(X), sd = tau)))
}
Create a container of class pomp
may <- pomp(data = data.frame(time = 1:100, Y = NA), times = "time",
rprocess = discrete.time.sim(step.fun = process.sim, delta.t = 1), rmeasure = measure.sim,
t0 = 0)
Parameters and an initial condition
theta <- c(r = 0.75, K = 10, Q = 3, H = 1, a = 1.3, sigma = 0.2,
tau = 0.1, X.0 = 5)
Simulate
dat <- simulate(may, params = theta, obs = TRUE, states = TRUE)
library(reshape2)
library(ggplot2)
dt <- melt(dat)[3:5]
names(dt) = c("time", "value", "variable")
ggplot(dt, aes(time, value, lty = variable)) + geom_line()
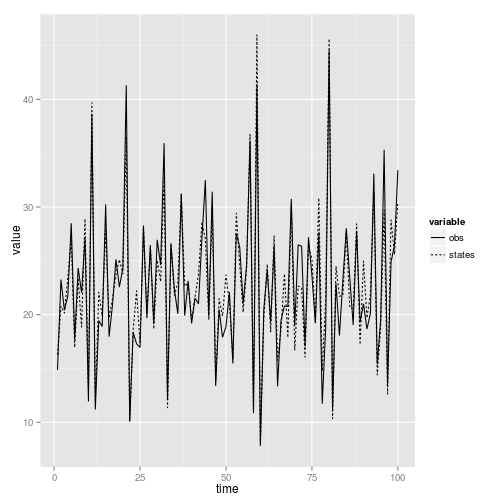
Note that simulate returns an instance of our object, with the data column filled in. This is not true if we give it arguments such as state or obs = TRUE as above. However, when we query for observed and true states in this way, the simulated data is not stored in our object. We must assign it with may <- pomp(data = ... where data is a data frame with columns "time" and "Y". This is much simpler if we use the default return:
may <- simulate(may, params = theta)
Measurement density
measure.density <- function(y, x, t, params, log, ...) {
tau <- params["tau"]
## state at time t:
X <- x["X"]
## observation at time t:
Y <- y["Y"]
## compute the likelihood of Y|X,tau
f <- dlnorm(x = Y, meanlog = log(X), sdlog = tau, log = log)
}
Add to our container
may <- pomp(may, dmeasure = measure.density)
Optionally, work in the transformed variable space instead by providing parameter and inverse parameter transforms.
may <- pomp(may, parameter.transform = function(params, ...) {
exp(params)
}, parameter.inv.transform = function(params, ...) {
log(params)
})
Run the particle filter. Note that this evaluates the likelihood of our simulated data (stored in the object) at the given (true) parameters.
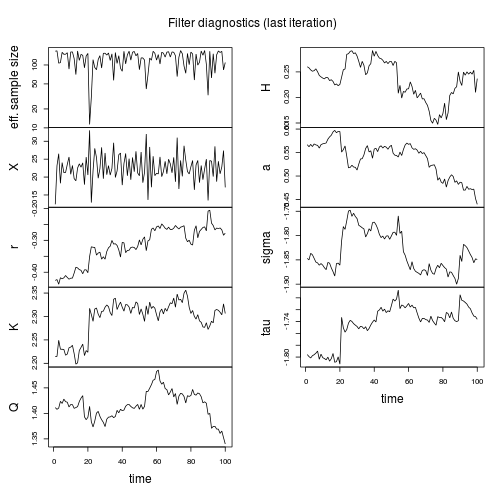
pf <- pfilter(may, params = theta, Np = 1000)
logLik(pf)
## [1] -304.7
Here we go, estimate the model by particle filter. This'll be slow...
theta.guess <- theta + c(abs(rnorm(length(theta)-1)),0)
estpars <- names(theta[1:7]) # which pars should be estimated. lets try and get them all
rw.sd <- theta[1:7]/(50*theta[1:7]) # a named vector of random walk values. This sets it to 0.02 for all pars
system.time(mf <-
mif(may, Nmif = 10, # Number of iterations
start = theta.guess, # Starting value of parameters
transform = TRUE, # work in transformed parameter space (e.g. logs avoid neg par values)
pars = estpars, # which parameters to estimate (others fixed at starting value)
rw.sd = rw.sd, # random walk width
Np = 200, # Number of particles
var.factor = 4, # scaling factor for random walk sd
ic.lag = 10, # fixed lag
cooling.factor = 0.999, # exponential cooling
max.fail = 10) # internal tolerance
)
## user system elapsed
## 22.018 0.008 22.106
compare.mif(mf)
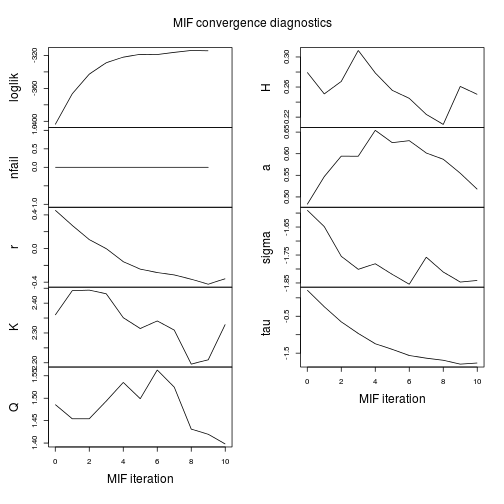
coef(mf)
## r K Q H a sigma tau X.0
## 0.6985 10.2655 4.0470 1.2844 1.6782 0.1587 0.1711 5.0000
logLik(mf)
## [1] -314.4
pf_est <- pfilter(may, params = theta.guess, Np = 1000)
logLik(pf_est) # initial loglik
## [1] -441.9
These functions apply to replicated mifs above.
theta.true <- coef(may)
theta.mif <- apply(sapply(mf, coef), 1, mean)
## Error: no method for coercing this S4 class to a vector
loglik.mif <- replicate(n = 10, logLik(pfilter(mf[[1]], params = theta.mif,
Np = 10000)))
## recover called non-interactively; frames dumped, use debugger() to view
## recover called non-interactively; frames dumped, use debugger() to view
## Error: error in evaluating the argument 'object' in selecting a method for
## function 'logLik': Error in pfilter(mf[[1]], params = theta.mif, Np =
## 10000) : error in evaluating the argument 'object' in selecting a method
## for function 'pfilter': Error in mf[[1]] : this S4 class is not
## subsettable
bl <- mean(loglik.mif)
## Error: object 'loglik.mif' not found
loglik.mif.est <- bl + log(mean(exp(loglik.mif - bl)))
## Error: object 'bl' not found
loglik.mif.se <- sd(exp(loglik.mif - bl))/exp(loglik.mif.est - bl)
## Error: object 'loglik.mif' not found
cboettig/nonparametric-bayes documentation built on May 13, 2019, 2:09 p.m.
library(pomp)
## Loading required package: mvtnorm
## Loading required package: subplex
## Loading required package: deSolve
opts_knit$set(upload.fun = socialR::flickr.url)
options(flickr_tags = "nonparametric-bayes")
process.sim <- function(x, t, params, delta.t, ...) {
# r = 0.75, K = 10, Q = 3, H = 1, a = 1.3
r <- params["r"]
K <- params["K"]
Q <- params["Q"]
H <- params["H"]
a <- params["a"]
sigma <- params["sigma"]
X <- x["X"]
epsilon <- exp(rlnorm(n = 1, mean = 0, sd = sigma))
c(X = unname(X * exp(r * delta.t * (1 - X/K) - a * X^(Q - 1)/(X^Q + H^Q)) *
epsilon))
}
measure.sim <- function(x, t, params, ...) {
tau <- params["tau"]
X <- x["X"]
y <- c(Y = unname(rlnorm(n = 1, meanlog = log(X), sd = tau)))
}
Create a container of class pomp
may <- pomp(data = data.frame(time = 1:100, Y = NA), times = "time",
rprocess = discrete.time.sim(step.fun = process.sim, delta.t = 1), rmeasure = measure.sim,
t0 = 0)
Parameters and an initial condition
theta <- c(r = 0.75, K = 10, Q = 3, H = 1, a = 1.3, sigma = 0.2,
tau = 0.1, X.0 = 5)
Simulate
dat <- simulate(may, params = theta, obs = TRUE, states = TRUE)
library(reshape2)
library(ggplot2)
dt <- melt(dat)[3:5]
names(dt) = c("time", "value", "variable")
ggplot(dt, aes(time, value, lty = variable)) + geom_line()

Note that simulate returns an instance of our object, with the data column filled in. This is not true if we give it arguments such as state or obs = TRUE as above. However, when we query for observed and true states in this way, the simulated data is not stored in our object. We must assign it with may <- pomp(data = ... where data is a data frame with columns "time" and "Y". This is much simpler if we use the default return:
may <- simulate(may, params = theta)
Measurement density
measure.density <- function(y, x, t, params, log, ...) {
tau <- params["tau"]
## state at time t:
X <- x["X"]
## observation at time t:
Y <- y["Y"]
## compute the likelihood of Y|X,tau
f <- dlnorm(x = Y, meanlog = log(X), sdlog = tau, log = log)
}
Add to our container
may <- pomp(may, dmeasure = measure.density)
Optionally, work in the transformed variable space instead by providing parameter and inverse parameter transforms.
may <- pomp(may, parameter.transform = function(params, ...) {
exp(params)
}, parameter.inv.transform = function(params, ...) {
log(params)
})
Run the particle filter. Note that this evaluates the likelihood of our simulated data (stored in the object) at the given (true) parameters.
pf <- pfilter(may, params = theta, Np = 1000)
logLik(pf)
## [1] -304.7
Here we go, estimate the model by particle filter. This'll be slow...
theta.guess <- theta + c(abs(rnorm(length(theta)-1)),0)
estpars <- names(theta[1:7]) # which pars should be estimated. lets try and get them all
rw.sd <- theta[1:7]/(50*theta[1:7]) # a named vector of random walk values. This sets it to 0.02 for all pars
system.time(mf <-
mif(may, Nmif = 10, # Number of iterations
start = theta.guess, # Starting value of parameters
transform = TRUE, # work in transformed parameter space (e.g. logs avoid neg par values)
pars = estpars, # which parameters to estimate (others fixed at starting value)
rw.sd = rw.sd, # random walk width
Np = 200, # Number of particles
var.factor = 4, # scaling factor for random walk sd
ic.lag = 10, # fixed lag
cooling.factor = 0.999, # exponential cooling
max.fail = 10) # internal tolerance
)
## user system elapsed
## 22.018 0.008 22.106
compare.mif(mf)


coef(mf)
## r K Q H a sigma tau X.0
## 0.6985 10.2655 4.0470 1.2844 1.6782 0.1587 0.1711 5.0000
logLik(mf)
## [1] -314.4
pf_est <- pfilter(may, params = theta.guess, Np = 1000)
logLik(pf_est) # initial loglik
## [1] -441.9
These functions apply to replicated mifs above.
theta.true <- coef(may)
theta.mif <- apply(sapply(mf, coef), 1, mean)
## Error: no method for coercing this S4 class to a vector
loglik.mif <- replicate(n = 10, logLik(pfilter(mf[[1]], params = theta.mif,
Np = 10000)))
## recover called non-interactively; frames dumped, use debugger() to view
## recover called non-interactively; frames dumped, use debugger() to view
## Error: error in evaluating the argument 'object' in selecting a method for
## function 'logLik': Error in pfilter(mf[[1]], params = theta.mif, Np =
## 10000) : error in evaluating the argument 'object' in selecting a method
## for function 'pfilter': Error in mf[[1]] : this S4 class is not
## subsettable
bl <- mean(loglik.mif)
## Error: object 'loglik.mif' not found
loglik.mif.est <- bl + log(mean(exp(loglik.mif - bl)))
## Error: object 'bl' not found
loglik.mif.se <- sd(exp(loglik.mif - bl))/exp(loglik.mif.est - bl)
## Error: object 'loglik.mif' not found
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
