inst/examples/semi-analytic-posteriors.md
In cboettig/nonparametric-bayes: Nonparametric Bayes inference for ecological models
Semi-analytic posteriors
Generating Model and parameters
Ricker model, parameterized as
$$X_{t+1} = X_t r e^{-\beta X_t + \sigma Z_t}$$
for random unit normal $Z_t$
f <- function(x,h,p) x * p[1] * exp(-x * p[2])
p <- c(exp(1), 1/10)
K <- 10 # approx, a li'l' less
Xo <- 1 # approx, a li'l' less
sigma_g <- 0.1
z_g <- function() rlnorm(1,0, sigma_g)
x_grid <- seq(0, 1.5 * K, length=50)
N <- 40
set.seed(123)
Sample Data
x <- numeric(N)
x[1] <- Xo
for(t in 1:(N-1))
x[t+1] = z_g() * f(x[t], h=0, p=p)
qplot(1:N, x)

Compute the posterior after marginalizing over $r$ and $\sigma$ parameters:
$$P(\beta | X) $$
Mt <- function(t, beta) log(x[t+1]) - log(x[t]) + beta * x[t]
beta_grid = seq(1e-5, 2, by=1e-3)
P_B.X <- sapply(beta_grid, function(beta){
Mt_vec = sapply(1:(N-1), Mt, beta)
sum_of_squares <- sum(Mt_vec^2)
square_of_sums <- sum(Mt_vec)^2
0.5 ^ (N/2-1) * (sum_of_squares - square_of_sums/(N-1)) ^ (N/2-1) / gamma(N/2-1)
})
qplot(beta_grid, -log(P_B.X))
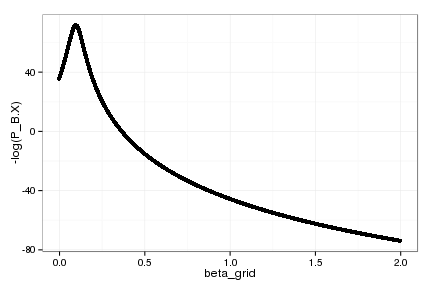
beta_grid[which.min(P_B.X)]
[1] 0.09801
Estimating the Allen model:
$$X_{t+1} = X_t \exp\left(r \left(1 - \frac{X_t}{K} \right\left(\frac{X - C}{K}\right)\right) $$
First re-parameterize so we can isolate an additive parameter, using standard quadratic form,
$$X_{t+1} = X_t \exp(c + b X_t + a X_t^2) $$
Where $c = \tfrac{-rC}{K}$, $b = \tfrac{r}{K}\left(\tfrac{C}{K} + 1\right)$, and $a = \tfrac{r}{K^2}$. Then after integrating out $c$ and $\sigma$ we have
Mt <- function(t, a, b) log(x[t+1]) - log(x[t]) - b * x[t] + a * x[t] ^ 2
Choosing a sensible grid is a bit more tricky following the transformation, particularly $b$ can be negative or positive, and much larger in magnitude than any of the original parameters. $a$ on the other hand, can be reasonably restricted:
b_grid = seq(0, .1, length=100)
a_grid = seq(0, .1, length=100)
We define the probability as a function of the remaining two parameters,
prob <- function(a, b){
Mt_vec = sapply(1:(N-1), Mt, a, b)
sum_of_squares <- sum(Mt_vec^2)
square_of_sums <- sum(Mt_vec)^2
0.5 ^ (N/2-1) * (sum_of_squares - square_of_sums/(N-1)) ^ (N/2-1) / gamma(N/2-1)
}
and loop over the grid on each
P_a_b.X <- sapply(a_grid, function(a)
sapply(b_grid, function(b) prob(a, b)))
We summarize with a contour plot
df = melt(P_a_b.X)
names(df) = c("a", "b", "lik")
ggplot(df, aes(a_grid[a], b_grid[b], z=-log(lik))) +
stat_contour(aes(color=..level..), binwidth=1) +
coord_cartesian(ylim=c(0, .025), xlim=c(0,.05))

Estimating the Myers model on this data:
$$X_{t+1} = Z_t \frac{r X_t^{\theta}}{1 + X_t^{\theta} / K}$$
With $Z_t$ lognormal, unit mean, std $\sigma$.
Mt <- function(t, theta, K) log(x[t+1]) - theta * log(x[t]) + log(1 + x[t] ^ theta / K)
theta_grid = seq(1e-5, 5, length=100)
K_grid = seq(1e-5, 30, length=100)
prob <- function(theta, K){
Mt_vec = sapply(1:(N-1), Mt, theta, K)
sum_of_squares <- sum(Mt_vec^2)
square_of_sums <- sum(Mt_vec)^2
0.5 ^ (N/2-1) * (sum_of_squares - square_of_sums/(N-1)) ^ (N/2-1) / gamma(N/2-1)
}
P_theta_K.X <- sapply(theta_grid, function(theta)
sapply(K_grid, function(k) prob(theta, k)))
require(reshape2)
df = melt(P_theta_K.X)
names(df) = c("theta", "K", "lik")
ggplot(df, aes(theta_grid[theta], K_grid[K], z=-log(lik))) + stat_contour(aes(color=..level..), binwidth=3)

cboettig/nonparametric-bayes documentation built on May 13, 2019, 2:09 p.m.
Semi-analytic posteriors
Generating Model and parameters
Ricker model, parameterized as
$$X_{t+1} = X_t r e^{-\beta X_t + \sigma Z_t}$$
for random unit normal $Z_t$
f <- function(x,h,p) x * p[1] * exp(-x * p[2])
p <- c(exp(1), 1/10)
K <- 10 # approx, a li'l' less
Xo <- 1 # approx, a li'l' less
sigma_g <- 0.1
z_g <- function() rlnorm(1,0, sigma_g)
x_grid <- seq(0, 1.5 * K, length=50)
N <- 40
set.seed(123)
Sample Data
x <- numeric(N)
x[1] <- Xo
for(t in 1:(N-1))
x[t+1] = z_g() * f(x[t], h=0, p=p)
qplot(1:N, x)

Compute the posterior after marginalizing over $r$ and $\sigma$ parameters:
$$P(\beta | X) $$
Mt <- function(t, beta) log(x[t+1]) - log(x[t]) + beta * x[t]
beta_grid = seq(1e-5, 2, by=1e-3)
P_B.X <- sapply(beta_grid, function(beta){
Mt_vec = sapply(1:(N-1), Mt, beta)
sum_of_squares <- sum(Mt_vec^2)
square_of_sums <- sum(Mt_vec)^2
0.5 ^ (N/2-1) * (sum_of_squares - square_of_sums/(N-1)) ^ (N/2-1) / gamma(N/2-1)
})
qplot(beta_grid, -log(P_B.X))

beta_grid[which.min(P_B.X)]
[1] 0.09801
Estimating the Allen model:
$$X_{t+1} = X_t \exp\left(r \left(1 - \frac{X_t}{K} \right\left(\frac{X - C}{K}\right)\right) $$
First re-parameterize so we can isolate an additive parameter, using standard quadratic form,
$$X_{t+1} = X_t \exp(c + b X_t + a X_t^2) $$
Where $c = \tfrac{-rC}{K}$, $b = \tfrac{r}{K}\left(\tfrac{C}{K} + 1\right)$, and $a = \tfrac{r}{K^2}$. Then after integrating out $c$ and $\sigma$ we have
Mt <- function(t, a, b) log(x[t+1]) - log(x[t]) - b * x[t] + a * x[t] ^ 2
Choosing a sensible grid is a bit more tricky following the transformation, particularly $b$ can be negative or positive, and much larger in magnitude than any of the original parameters. $a$ on the other hand, can be reasonably restricted:
b_grid = seq(0, .1, length=100)
a_grid = seq(0, .1, length=100)
We define the probability as a function of the remaining two parameters,
prob <- function(a, b){
Mt_vec = sapply(1:(N-1), Mt, a, b)
sum_of_squares <- sum(Mt_vec^2)
square_of_sums <- sum(Mt_vec)^2
0.5 ^ (N/2-1) * (sum_of_squares - square_of_sums/(N-1)) ^ (N/2-1) / gamma(N/2-1)
}
and loop over the grid on each
P_a_b.X <- sapply(a_grid, function(a)
sapply(b_grid, function(b) prob(a, b)))
We summarize with a contour plot
df = melt(P_a_b.X)
names(df) = c("a", "b", "lik")
ggplot(df, aes(a_grid[a], b_grid[b], z=-log(lik))) +
stat_contour(aes(color=..level..), binwidth=1) +
coord_cartesian(ylim=c(0, .025), xlim=c(0,.05))

Estimating the Myers model on this data:
$$X_{t+1} = Z_t \frac{r X_t^{\theta}}{1 + X_t^{\theta} / K}$$
With $Z_t$ lognormal, unit mean, std $\sigma$.
Mt <- function(t, theta, K) log(x[t+1]) - theta * log(x[t]) + log(1 + x[t] ^ theta / K)
theta_grid = seq(1e-5, 5, length=100)
K_grid = seq(1e-5, 30, length=100)
prob <- function(theta, K){
Mt_vec = sapply(1:(N-1), Mt, theta, K)
sum_of_squares <- sum(Mt_vec^2)
square_of_sums <- sum(Mt_vec)^2
0.5 ^ (N/2-1) * (sum_of_squares - square_of_sums/(N-1)) ^ (N/2-1) / gamma(N/2-1)
}
P_theta_K.X <- sapply(theta_grid, function(theta)
sapply(K_grid, function(k) prob(theta, k)))
require(reshape2)
df = melt(P_theta_K.X)
names(df) = c("theta", "K", "lik")
ggplot(df, aes(theta_grid[theta], K_grid[K], z=-log(lik))) + stat_contour(aes(color=..level..), binwidth=3)

Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
