In marionlouveaux/cellviz3d: Tools for 3D/4D interactive visualization of cells and biological tissues
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>",
fig.path = "man/figures/README-",
out.width = "100%"
)
cellviz3d
The goal of {cellviz3d} is to propose visualisation of meshes created from the segmentation of 3D/4D microscopic images, and point clouds. In particular, it helps visualising meshes from MorphographX, in combination with {mgx2r}, or point clouds from the MaMuT Fiji plugin, in combination with {mamut2r}.
How to cite
To cite {cellviz3d}, call the R built-in command citation("cellviz3d").
Marion Louveaux. (2018, October 19). cellviz3d: a R package for the 3D and 4D interactive visualization of cells and biological tissues (Version v0.0.2). Zenodo. http://doi.org/10.5281/zenodo.1467416
Installation
You can install the released version of {cellviz3d} from Github with:
# install.packages("devtools")
devtools::install_github("marionlouveaux/cellviz3d")
# With vignettes
devtools::install_github("marionlouveaux/cellviz3d", build_vignettes = TRUE)
Full documentation with {pkgdown}
See full documentation created with {pkgdown} at https://marionlouveaux.github.io/cellviz3d/
Vignette
A vignette is available in the package if you built it during installation with build_vignettes = TRUE:
vignette("cellviz3d-mgx_meshes", package = "cellviz3d"): how to build 3d interactive plots of meshes as issued from {mgx2r}
Example
The data in the example below were created with the package {mgx2r}, on the example dataset of {mgx2r}. This dataset is a timelapse recording of the development of a shoot apical meristem of the plant \emph{Arabidopsis thaliana} expressing a membrane marker. I took one 3D stack every 12h and have 5 timepoints in total. To know more about the example dataset of {mgx2r}, see help.search("mgx2r-package").
library(cellviz3d)
library(plotly)
# Read data
myMesh <- readRDS(system.file("extdata",
"mgx/mesh_meristem_full_T0.rds",
package = "cellviz3d"))
myCellGraph <- readRDS(system.file("extdata",
"mgx/cellGraph_meristem_full_T0.rds",
package = "cellviz3d"))
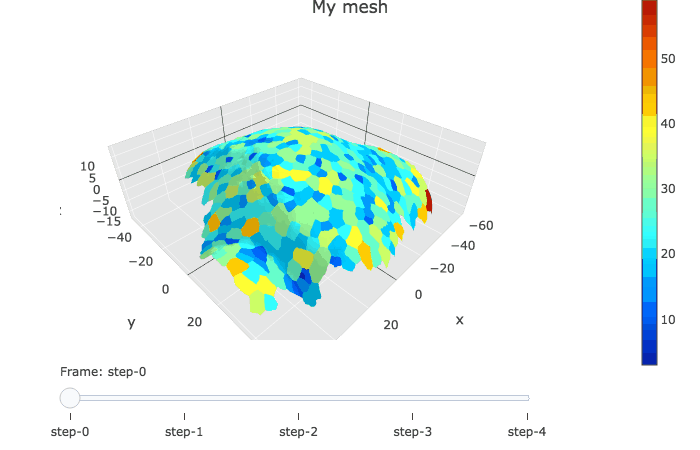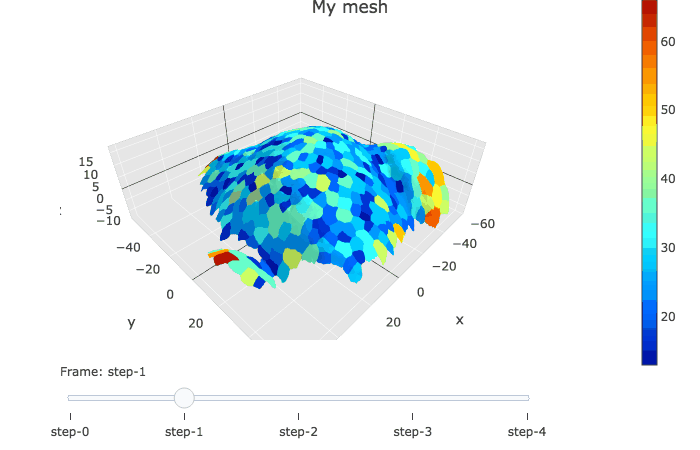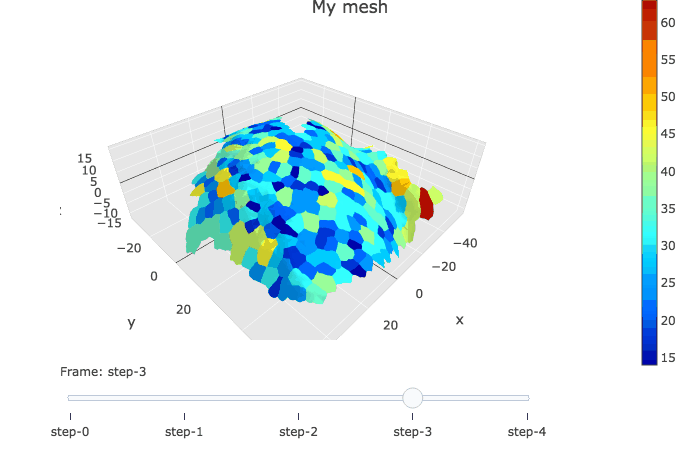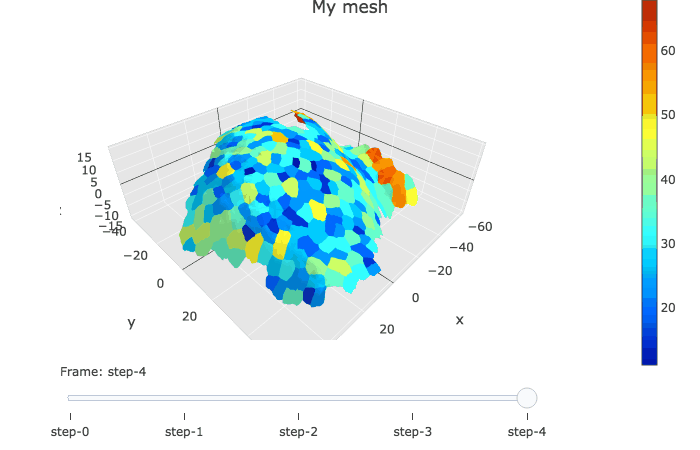
plotlyMesh() creates a plotly graph of type mesh 3D, with custom colors and custom hover information for a single mesh. Below, the mesh is displayed with one color and one cell label per biological cell. Cell label is visible when hovering over the cell center.
meshCellcenter <- myCellGraph$vertices[,c("label","x", "y", "z")]
plotlyMesh(meshExample = myMesh,
meshColors = myMesh$allColors$Col_label,
meshCellcenter = meshCellcenter) %>%
layout(scene = list(aspectmode = "data"))
knitr::include_graphics("https://raw.githubusercontent.com/marionlouveaux/cellviz3d/master/inst/extdata/mgx/img/p1labels.png")
For more examples, see the vignette of {cellviz3d} and {mgx2r}.
Acknowledgements
Many thanks to @friep for her help on the plotlySpots_all() function.
Code of conduct
Please note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.
marionlouveaux/cellviz3d documentation built on May 20, 2019, 9:55 a.m.
knitr::opts_chunk$set( collapse = TRUE, comment = "#>", fig.path = "man/figures/README-", out.width = "100%" )
cellviz3d
The goal of {cellviz3d} is to propose visualisation of meshes created from the segmentation of 3D/4D microscopic images, and point clouds. In particular, it helps visualising meshes from MorphographX, in combination with {mgx2r}, or point clouds from the MaMuT Fiji plugin, in combination with {mamut2r}.
How to cite
To cite {cellviz3d}, call the R built-in command citation("cellviz3d").
Marion Louveaux. (2018, October 19). cellviz3d: a R package for the 3D and 4D interactive visualization of cells and biological tissues (Version v0.0.2). Zenodo. http://doi.org/10.5281/zenodo.1467416
Installation
You can install the released version of {cellviz3d} from Github with:
# install.packages("devtools") devtools::install_github("marionlouveaux/cellviz3d") # With vignettes devtools::install_github("marionlouveaux/cellviz3d", build_vignettes = TRUE)
Full documentation with {pkgdown}
See full documentation created with {pkgdown} at https://marionlouveaux.github.io/cellviz3d/
Vignette
A vignette is available in the package if you built it during installation with build_vignettes = TRUE:
vignette("cellviz3d-mgx_meshes", package = "cellviz3d"): how to build 3d interactive plots of meshes as issued from {mgx2r}
Example
The data in the example below were created with the package {mgx2r}, on the example dataset of {mgx2r}. This dataset is a timelapse recording of the development of a shoot apical meristem of the plant \emph{Arabidopsis thaliana} expressing a membrane marker. I took one 3D stack every 12h and have 5 timepoints in total. To know more about the example dataset of {mgx2r}, see help.search("mgx2r-package").
library(cellviz3d) library(plotly)
# Read data myMesh <- readRDS(system.file("extdata", "mgx/mesh_meristem_full_T0.rds", package = "cellviz3d")) myCellGraph <- readRDS(system.file("extdata", "mgx/cellGraph_meristem_full_T0.rds", package = "cellviz3d"))
plotlyMesh() creates a plotly graph of type mesh 3D, with custom colors and custom hover information for a single mesh. Below, the mesh is displayed with one color and one cell label per biological cell. Cell label is visible when hovering over the cell center.
meshCellcenter <- myCellGraph$vertices[,c("label","x", "y", "z")] plotlyMesh(meshExample = myMesh, meshColors = myMesh$allColors$Col_label, meshCellcenter = meshCellcenter) %>% layout(scene = list(aspectmode = "data"))
knitr::include_graphics("https://raw.githubusercontent.com/marionlouveaux/cellviz3d/master/inst/extdata/mgx/img/p1labels.png")

For more examples, see the vignette of {cellviz3d} and {mgx2r}.
Acknowledgements
Many thanks to @friep for her help on the plotlySpots_all() function.
Code of conduct
Please note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
