In natverse/neuronbridger: R client utilities for interacting with the NeuronBridge matching service
knitr::opts_chunk$set(
collapse = TRUE,
comment = "#>"
)
Designing a new split-GAL4 line
In this guide, we start with a neuron from the hemibrain connectome and use NeuronBridge to design a potential split-GAL4 line.
Let us say we are interested in the hemibrain neuron LAL051. This is a local neuron in a lower part of the fly's brain called the 'LAL' that synapses strongly onto 'descending neurons' that guide motor control for the fly.
We might want to generate a 'sparse' genetic driver line that more or less just 'targets' this neuron, allowing us to express transgenes such as flourescent proteins, calcium sensitive molecules for imaging or optogenetic tools just in this cell type. This can be very very useful for many experimental purposes.
Getting our target EM neuron in 3D
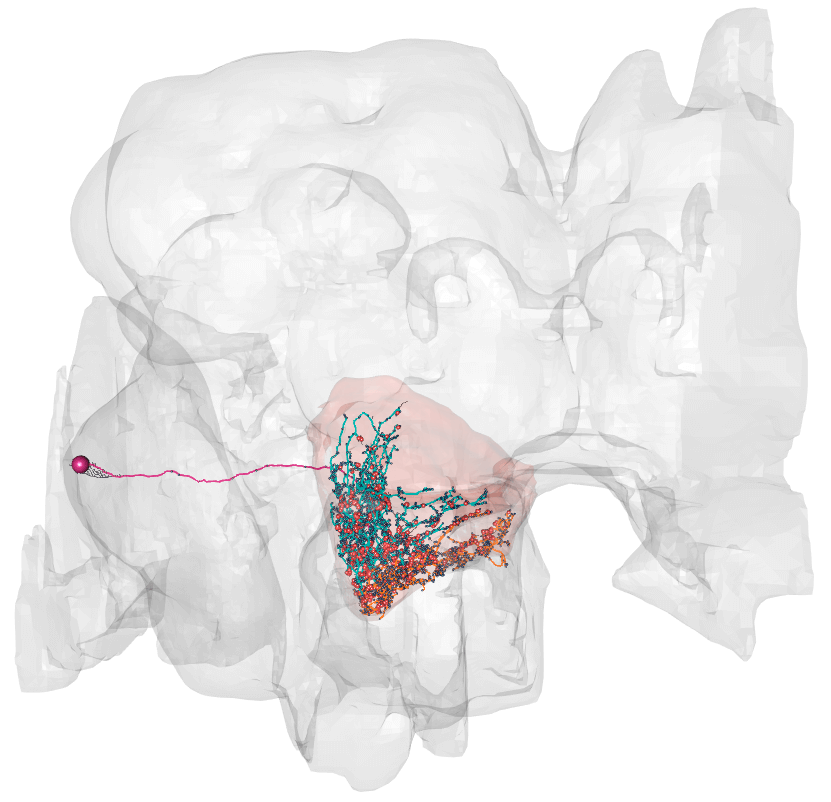
We can use the package hemibrainr to see our target neuron, and how it break down into axon and dendrite:
# Load a package we will need
library(hemibrainr) # to get this to work, please see:
# Find out neuron of interest
LAL051.info = neuprint_search("LAL051", field = "type")
LAL051.id = as.character(LAL051.info$bodyid)
LAL051.n = hemibrain_read_neurons(LAL051.id)
# Get the LAL
lal = neuprint_ROI_mesh("LAL(R)")
# Plot this neuron
nat::nopen3d()
plot3d(hemibrain.surf, col = "grey", alpha = 0.1)
plot3d(lal, col = hemibrain_bright_colors["red"], alpha = 0.1, add = TRUE)
hemibrain_view()
plot3d_split(LAL051.n, lwd = 2, soma = 400)
### Dendrite in blue, axon in orange, outouts in red, inputs in navy blue
Getting our target neuron as a MIP
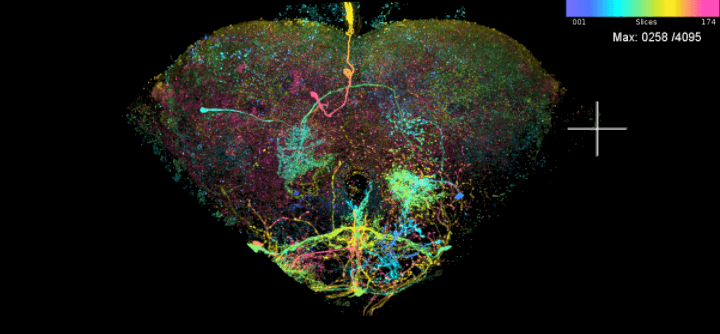
We can now plot this neuron as a maximal projection image (MIP). This is a 2D image where colour encodes depth. Images liek this for all hemibrain neurons have been used to find matches amoung light-level expression data (Otsuna et al. 2018).
# What do we have on it?
nb.info = neuronbridge_info(LAL051.id)
View(nb.info)
# Let us now see the related MIP file
LAL051.mip = neuronbridge_mip(LAL051.id)
## This gets every MIP file associated with id
# Plot the MIP image in an rgl viewer
clear3d()
plot_mip(LAL051.mip)

Search for lines that contain target neuron
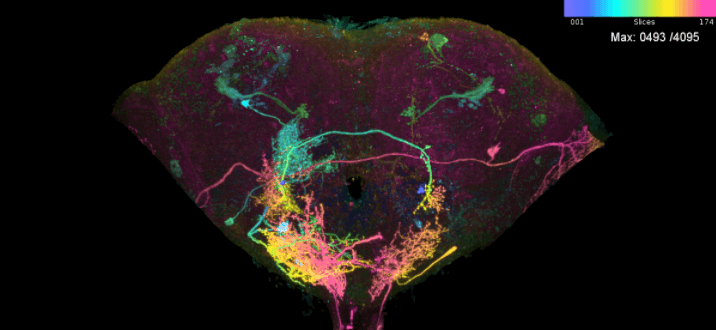
We can get MIP images for rthe potential matches (genetic drive lines, mostly GAL4).
# Run search, looked at precomputed data
ns.s = neuronbridge_search(LAL051.id, threshold = 2000)
# scan hits
open3d()
scan_mip(mips = ns.s, no.hits = nrow(ns.s))
The best matches seem to be, just: R58G03, R71D08
Design split
# Based on the results above
line1 = "R58G03"
line2 = "R71D08"
# What neurons might we expect?
potential.split = neuronbridge_predict_split(line1 = line1, line2 = line2,
threshold1 = 2000, threshold2 = 2000)
message("Potentially ~", nrow(potential.split), " neurons in this predicted split")
View(potential.split)
natverse/neuronbridger documentation built on Nov. 12, 2023, 11:21 a.m.
knitr::opts_chunk$set( collapse = TRUE, comment = "#>" )
Designing a new split-GAL4 line
In this guide, we start with a neuron from the hemibrain connectome and use NeuronBridge to design a potential split-GAL4 line.
Let us say we are interested in the hemibrain neuron LAL051. This is a local neuron in a lower part of the fly's brain called the 'LAL' that synapses strongly onto 'descending neurons' that guide motor control for the fly.
We might want to generate a 'sparse' genetic driver line that more or less just 'targets' this neuron, allowing us to express transgenes such as flourescent proteins, calcium sensitive molecules for imaging or optogenetic tools just in this cell type. This can be very very useful for many experimental purposes.
Getting our target EM neuron in 3D
We can use the package hemibrainr to see our target neuron, and how it break down into axon and dendrite:
# Load a package we will need library(hemibrainr) # to get this to work, please see: # Find out neuron of interest LAL051.info = neuprint_search("LAL051", field = "type") LAL051.id = as.character(LAL051.info$bodyid) LAL051.n = hemibrain_read_neurons(LAL051.id) # Get the LAL lal = neuprint_ROI_mesh("LAL(R)") # Plot this neuron nat::nopen3d() plot3d(hemibrain.surf, col = "grey", alpha = 0.1) plot3d(lal, col = hemibrain_bright_colors["red"], alpha = 0.1, add = TRUE) hemibrain_view() plot3d_split(LAL051.n, lwd = 2, soma = 400) ### Dendrite in blue, axon in orange, outouts in red, inputs in navy blue

Getting our target neuron as a MIP
We can now plot this neuron as a maximal projection image (MIP). This is a 2D image where colour encodes depth. Images liek this for all hemibrain neurons have been used to find matches amoung light-level expression data (Otsuna et al. 2018).
# What do we have on it? nb.info = neuronbridge_info(LAL051.id) View(nb.info) # Let us now see the related MIP file LAL051.mip = neuronbridge_mip(LAL051.id) ## This gets every MIP file associated with id # Plot the MIP image in an rgl viewer clear3d() plot_mip(LAL051.mip)

Search for lines that contain target neuron
We can get MIP images for rthe potential matches (genetic drive lines, mostly GAL4).
# Run search, looked at precomputed data ns.s = neuronbridge_search(LAL051.id, threshold = 2000) # scan hits open3d() scan_mip(mips = ns.s, no.hits = nrow(ns.s))
The best matches seem to be, just: R58G03, R71D08


Design split
# Based on the results above line1 = "R58G03" line2 = "R71D08" # What neurons might we expect? potential.split = neuronbridge_predict_split(line1 = line1, line2 = line2, threshold1 = 2000, threshold2 = 2000) message("Potentially ~", nrow(potential.split), " neurons in this predicted split") View(potential.split)
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
