README.md
In olascodgreat/samife: A Grammar of Data Manipulation
dplyr 
Overview
dplyr is a grammar of data manipulation, providing a consistent set of
verbs that help you solve the most common data manipulation challenges:
mutate() adds new variables that are functions of existing
variablesselect() picks variables based on their names.filter() picks cases based on their values.summarise() reduces multiple values down to a single summary.arrange() changes the ordering of the rows.
These all combine naturally with group_by() which allows you to
perform any operation “by group”. You can learn more about them in
vignette("dplyr"). As well as these single-table verbs, dplyr also
provides a variety of two-table verbs, which you can learn about in
vignette("two-table").
dplyr is designed to abstract over how the data is stored. That means as
well as working with local data frames, you can also work with remote
database tables, using exactly the same R code. Install the dbplyr
package then read vignette("databases", package = "dbplyr").
If you are new to dplyr, the best place to start is the data import
chapter in R for data science.
Installation
# The easiest way to get dplyr is to install the whole tidyverse:
install.packages("tidyverse")
# Alternatively, install just dplyr:
install.packages("dplyr")
Development version
To get a bug fix, or use a feature from the development version, you can
install dplyr from GitHub.
# install.packages("devtools")
devtools::install_github("tidyverse/dplyr")
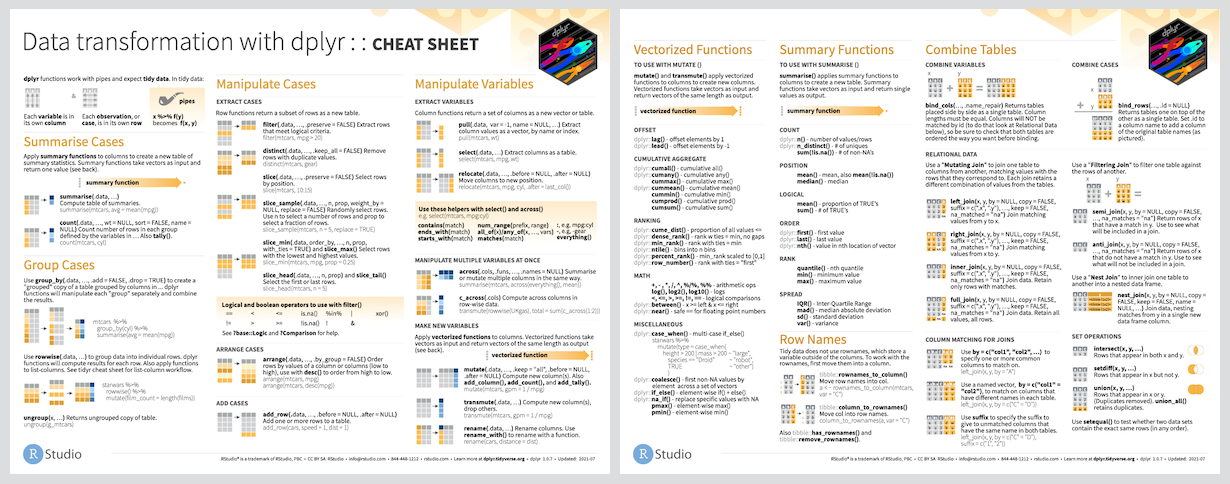
Cheatsheet
Usage
library(dplyr)
starwars %>%
filter(species == "Droid")
#> # A tibble: 5 x 13
#> name height mass hair_color skin_color eye_color birth_year gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr>
#> 1 C-3PO 167 75 <NA> gold yellow 112 <NA>
#> 2 R2-D2 96 32 <NA> white, bl… red 33 <NA>
#> 3 R5-D4 97 32 <NA> white, red red NA <NA>
#> 4 IG-88 200 140 none metal red 15 none
#> 5 BB8 NA NA none none black NA none
#> # ... with 5 more variables: homeworld <chr>, species <chr>, films <list>,
#> # vehicles <list>, starships <list>
starwars %>%
select(name, ends_with("color"))
#> # A tibble: 87 x 4
#> name hair_color skin_color eye_color
#> <chr> <chr> <chr> <chr>
#> 1 Luke Skywalker blond fair blue
#> 2 C-3PO <NA> gold yellow
#> 3 R2-D2 <NA> white, blue red
#> 4 Darth Vader none white yellow
#> 5 Leia Organa brown light brown
#> # ... with 82 more rows
starwars %>%
mutate(name, bmi = mass / ((height / 100) ^ 2)) %>%
select(name:mass, bmi)
#> # A tibble: 87 x 4
#> name height mass bmi
#> <chr> <int> <dbl> <dbl>
#> 1 Luke Skywalker 172 77 26.0
#> 2 C-3PO 167 75 26.9
#> 3 R2-D2 96 32 34.7
#> 4 Darth Vader 202 136 33.3
#> 5 Leia Organa 150 49 21.8
#> # ... with 82 more rows
starwars %>%
arrange(desc(mass))
#> # A tibble: 87 x 13
#> name height mass hair_color skin_color eye_color birth_year gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr>
#> 1 Jabb… 175 1358 <NA> green-tan… orange 600 herma…
#> 2 Grie… 216 159 none brown, wh… green, y… NA male
#> 3 IG-88 200 140 none metal red 15 none
#> 4 Dart… 202 136 none white yellow 41.9 male
#> 5 Tarf… 234 136 brown brown blue NA male
#> # ... with 82 more rows, and 5 more variables: homeworld <chr>,
#> # species <chr>, films <list>, vehicles <list>, starships <list>
starwars %>%
group_by(species) %>%
summarise(
n = n(),
mass = mean(mass, na.rm = TRUE)
) %>%
filter(n > 1)
#> # A tibble: 9 x 3
#> species n mass
#> <chr> <int> <dbl>
#> 1 <NA> 5 48
#> 2 Droid 5 69.8
#> 3 Gungan 3 74
#> 4 Human 35 82.8
#> 5 Kaminoan 2 88
#> # ... with 4 more rows
Getting help
If you encounter a clear bug, please file a minimal reproducible example
on github. For questions
and other discussion, please use
community.rstudio.com, or the
manipulatr mailing list.
Please note that this project is released with a Contributor Code of
Conduct. By participating in this project
you agree to abide by its terms.
olascodgreat/samife documentation built on May 13, 2019, 6:11 p.m.
dplyr 
Overview
dplyr is a grammar of data manipulation, providing a consistent set of verbs that help you solve the most common data manipulation challenges:
mutate()adds new variables that are functions of existing variablesselect()picks variables based on their names.filter()picks cases based on their values.summarise()reduces multiple values down to a single summary.arrange()changes the ordering of the rows.
These all combine naturally with group_by() which allows you to
perform any operation “by group”. You can learn more about them in
vignette("dplyr"). As well as these single-table verbs, dplyr also
provides a variety of two-table verbs, which you can learn about in
vignette("two-table").
dplyr is designed to abstract over how the data is stored. That means as
well as working with local data frames, you can also work with remote
database tables, using exactly the same R code. Install the dbplyr
package then read vignette("databases", package = "dbplyr").
If you are new to dplyr, the best place to start is the data import chapter in R for data science.
Installation
# The easiest way to get dplyr is to install the whole tidyverse:
install.packages("tidyverse")
# Alternatively, install just dplyr:
install.packages("dplyr")
Development version
To get a bug fix, or use a feature from the development version, you can install dplyr from GitHub.
# install.packages("devtools")
devtools::install_github("tidyverse/dplyr")
Cheatsheet
Usage
library(dplyr)
starwars %>%
filter(species == "Droid")
#> # A tibble: 5 x 13
#> name height mass hair_color skin_color eye_color birth_year gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr>
#> 1 C-3PO 167 75 <NA> gold yellow 112 <NA>
#> 2 R2-D2 96 32 <NA> white, bl… red 33 <NA>
#> 3 R5-D4 97 32 <NA> white, red red NA <NA>
#> 4 IG-88 200 140 none metal red 15 none
#> 5 BB8 NA NA none none black NA none
#> # ... with 5 more variables: homeworld <chr>, species <chr>, films <list>,
#> # vehicles <list>, starships <list>
starwars %>%
select(name, ends_with("color"))
#> # A tibble: 87 x 4
#> name hair_color skin_color eye_color
#> <chr> <chr> <chr> <chr>
#> 1 Luke Skywalker blond fair blue
#> 2 C-3PO <NA> gold yellow
#> 3 R2-D2 <NA> white, blue red
#> 4 Darth Vader none white yellow
#> 5 Leia Organa brown light brown
#> # ... with 82 more rows
starwars %>%
mutate(name, bmi = mass / ((height / 100) ^ 2)) %>%
select(name:mass, bmi)
#> # A tibble: 87 x 4
#> name height mass bmi
#> <chr> <int> <dbl> <dbl>
#> 1 Luke Skywalker 172 77 26.0
#> 2 C-3PO 167 75 26.9
#> 3 R2-D2 96 32 34.7
#> 4 Darth Vader 202 136 33.3
#> 5 Leia Organa 150 49 21.8
#> # ... with 82 more rows
starwars %>%
arrange(desc(mass))
#> # A tibble: 87 x 13
#> name height mass hair_color skin_color eye_color birth_year gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr>
#> 1 Jabb… 175 1358 <NA> green-tan… orange 600 herma…
#> 2 Grie… 216 159 none brown, wh… green, y… NA male
#> 3 IG-88 200 140 none metal red 15 none
#> 4 Dart… 202 136 none white yellow 41.9 male
#> 5 Tarf… 234 136 brown brown blue NA male
#> # ... with 82 more rows, and 5 more variables: homeworld <chr>,
#> # species <chr>, films <list>, vehicles <list>, starships <list>
starwars %>%
group_by(species) %>%
summarise(
n = n(),
mass = mean(mass, na.rm = TRUE)
) %>%
filter(n > 1)
#> # A tibble: 9 x 3
#> species n mass
#> <chr> <int> <dbl>
#> 1 <NA> 5 48
#> 2 Droid 5 69.8
#> 3 Gungan 3 74
#> 4 Human 35 82.8
#> 5 Kaminoan 2 88
#> # ... with 4 more rows
Getting help
If you encounter a clear bug, please file a minimal reproducible example on github. For questions and other discussion, please use community.rstudio.com, or the manipulatr mailing list.
Please note that this project is released with a Contributor Code of Conduct. By participating in this project you agree to abide by its terms.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.