README.md
In systemPipeR/systemPipeShiny: systemPipeShiny: An Interactive Framework for Workflow Management and Visualization
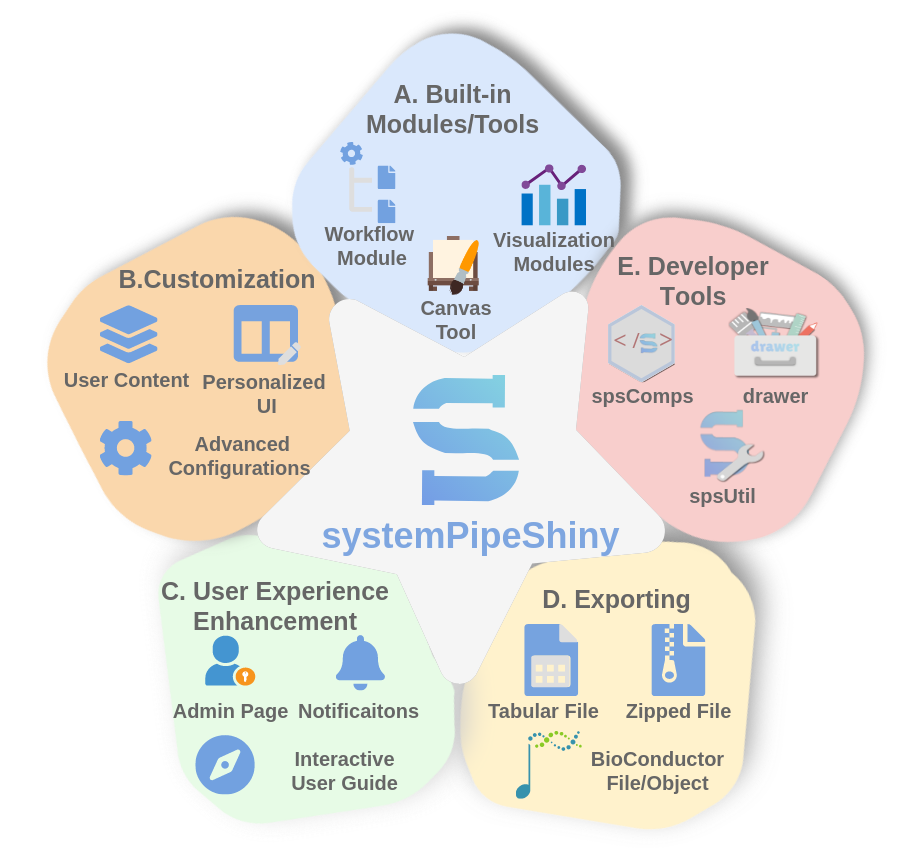
systemPipeShiny 

systemPipeShiny(SPS) a Shiny-based R/Bioconductor package that extends the widely used
systemPipeR workflow
environment with data visualization and a versatile graphical user interface.
SPS can work as a general framework to build custom web apps on data analysis and visualization.
Besides, SPS provides many developer tools that are distributed as separate packages.
4 minutes quick overview
You need to unmuted it to hear the sound.
Demos
SPS has provided a variety of options to change how it work. Here are some examples.
| Type and link| option changed | notes |
| --- | --- | --- |
| Default full installation | See installation | full app, may take longer (~15s) to load |
| Minimum installation | See installation | no modules installed |
| Login enabled | login_screen = TRUE; login_theme = "empty" | no modules installed |
| Login and login themes | login_screen = TRUE; login_theme = "random" | no modules installed |
| App admin page | admin_page = TRUE | use the link or simply add "?admin" to the end of URL of any demos |
For the login required demos, the app account name is "user" password "user".
For the admin login, account name "admin", password "admin".
Please DO NOT delete or change password when you are trying the admin features.
Although shinyapps.io will reset the app once a while, this will affect other people
who are viewing the demo simultaneously.
Rstudio Cloud
There is an Rstudio Cloud project instance
that you can also play with. You need to create a free new account. Two Bioconductor
related modules - workflow & RNAseq are not installed. They require more than 1GB
RAM to install and to run which is beyond the limit of a free account.
Docker
Use systempipe_docker for instructions.
Documents
To see all the details of SPS, read the user manual on our website.
Installation
Full
if (!requireNamespace("BiocManager", quietly=TRUE))
install.packages("BiocManager")
BiocManager::install("systemPipeShiny", dependencies=TRUE)
This will install all required packages including suggested packages that
are required by the core modules. Be aware, it will take quite some time if you
are installing on Linux where only source installation is available. Windows and Mac
binary installations will be much faster.
Minimum
To install the package, please use the BiocManager::install command:
if (!requireNamespace("BiocManager", quietly=TRUE))
install.packages("BiocManager")
BiocManager::install("systemPipeShiny")
By the minimum installation, all the 3 core modules are not installed. You
can still start the app, and when you start the app and click on these modules,
it will tell to enable these modules, what packages to install and what
command you need to run.
Just follow the instructions. Install as you need.
Most recent
To obtain the most recent updates immediately, one can install it directly from
GitHub as follow:
if (!requireNamespace("remotes", quietly=TRUE))
install.packages("remotes")
remotes::install("systemPipeR/systemPipeShiny", dependencies=TRUE)
Similarly, remotes::install("systemPipeR/systemPipeShiny") for the minimum develop
version.
Linux
If you are on Linux, you may also need the following system dependencies before installing SPS.
Different distributions
may have different commands, but the following commands are examples for Ubuntu:
sudo apt-get install -y libicu-dev
sudo apt-get install -y pandoc
sudo apt-get install -y zlib1g-dev
sudo apt-get install -y libcurl4-openssl-dev
sudo apt-get install -y libssl-dev
sudo apt-get install -y make
Quick start
This is a basic example which shows how to use systempipeShiny package:
## Imports the library
library(systemPipeShiny)
## Creates the project directory
spsInit()
By default, a project folder is created and named as SPS_+DATE.
This project folder provides all the necessary files to launch the application.
If you are using Rstudio, global.R file will be opened automatically and this is
the only file you may need to make custom changes if there is any.
Click the green "Run App" button in Rstudio if you are on the global.R file or
run following in console to start the app.
## Launching the interface
shiny::runApp()
options
Change some of the options
listed in global.R will change how the app behave, for example, modules to load,
title, logo, whether to apply user authentication, and more ...
Other packages in systemPipeShiny
| Package | Description | Documents | Function reference | Demo |
| --- | --- | --- | :---: | --- |
| systemPipeShiny | SPS main package |website|link | demo|
|
systemPipeShiny | SPS main package |website|link | demo|
| spsComps | SPS UI and server components |website|link | demo|
|
spsComps | SPS UI and server components |website|link | demo|
| drawer | SPS interactive image editing tool |website|link | demo|
|
drawer | SPS interactive image editing tool |website|link | demo|
| spsUtil | SPS utility functions |website|link | NA|
spsUtil | SPS utility functions |website|link | NA|
More Tutorials
Expand
You need to unmuted it to hear the sound.
Workflow Module
RNAseq Module
Quick ggplot Module
Canvas Tool
Screenshots of SPS
Expand
Full app

Loading screens

Workflow module

Workflow Execution

RNASeq module


Canvas

Admin



Debugging

Contact & contributions
Please use https://github.com/systemPipeR/systemPipeShiny/issues for reporting bugs,
issues or for suggesting new features to be implemented.
We'd love to hear from all users and developers. Submit your pull request if you
have new thoughts or improvements.
systemPipeR/systemPipeShiny documentation built on Oct. 17, 2023, 3:40 a.m.
systemPipeShiny 
systemPipeShiny(SPS) a Shiny-based R/Bioconductor package that extends the widely used
systemPipeR workflow
environment with data visualization and a versatile graphical user interface.
SPS can work as a general framework to build custom web apps on data analysis and visualization.
Besides, SPS provides many developer tools that are distributed as separate packages.
4 minutes quick overview
You need to unmuted it to hear the sound.

Demos
SPS has provided a variety of options to change how it work. Here are some examples.
| Type and link| option changed | notes |
| --- | --- | --- |
| Default full installation | See installation | full app, may take longer (~15s) to load |
| Minimum installation | See installation | no modules installed |
| Login enabled | login_screen = TRUE; login_theme = "empty" | no modules installed |
| Login and login themes | login_screen = TRUE; login_theme = "random" | no modules installed |
| App admin page | admin_page = TRUE | use the link or simply add "?admin" to the end of URL of any demos |
For the login required demos, the app account name is "user" password "user".
For the admin login, account name "admin", password "admin".
Please DO NOT delete or change password when you are trying the admin features. Although shinyapps.io will reset the app once a while, this will affect other people who are viewing the demo simultaneously.
Rstudio Cloud
There is an Rstudio Cloud project instance that you can also play with. You need to create a free new account. Two Bioconductor related modules - workflow & RNAseq are not installed. They require more than 1GB RAM to install and to run which is beyond the limit of a free account.
Docker
Use systempipe_docker for instructions.
Documents
To see all the details of SPS, read the user manual on our website.
Installation
Full
if (!requireNamespace("BiocManager", quietly=TRUE))
install.packages("BiocManager")
BiocManager::install("systemPipeShiny", dependencies=TRUE)
This will install all required packages including suggested packages that are required by the core modules. Be aware, it will take quite some time if you are installing on Linux where only source installation is available. Windows and Mac binary installations will be much faster.
Minimum
To install the package, please use the BiocManager::install command:
if (!requireNamespace("BiocManager", quietly=TRUE))
install.packages("BiocManager")
BiocManager::install("systemPipeShiny")
By the minimum installation, all the 3 core modules are not installed. You can still start the app, and when you start the app and click on these modules, it will tell to enable these modules, what packages to install and what command you need to run. Just follow the instructions. Install as you need.
Most recent
To obtain the most recent updates immediately, one can install it directly from GitHub as follow:
if (!requireNamespace("remotes", quietly=TRUE))
install.packages("remotes")
remotes::install("systemPipeR/systemPipeShiny", dependencies=TRUE)
Similarly, remotes::install("systemPipeR/systemPipeShiny") for the minimum develop
version.
Linux
If you are on Linux, you may also need the following system dependencies before installing SPS. Different distributions may have different commands, but the following commands are examples for Ubuntu:
sudo apt-get install -y libicu-dev
sudo apt-get install -y pandoc
sudo apt-get install -y zlib1g-dev
sudo apt-get install -y libcurl4-openssl-dev
sudo apt-get install -y libssl-dev
sudo apt-get install -y make
Quick start
This is a basic example which shows how to use systempipeShiny package:
## Imports the library
library(systemPipeShiny)
## Creates the project directory
spsInit()
By default, a project folder is created and named as SPS_+DATE.
This project folder provides all the necessary files to launch the application.
If you are using Rstudio, global.R file will be opened automatically and this is
the only file you may need to make custom changes if there is any.
Click the green "Run App" button in Rstudio if you are on the global.R file or
run following in console to start the app.
## Launching the interface
shiny::runApp()
options
Change some of the options
listed in global.R will change how the app behave, for example, modules to load,
title, logo, whether to apply user authentication, and more ...
Other packages in systemPipeShiny
| Package | Description | Documents | Function reference | Demo |
| --- | --- | --- | :---: | --- |
|![]() systemPipeShiny | SPS main package |website|link | demo|
|
systemPipeShiny | SPS main package |website|link | demo|
|![]() spsComps | SPS UI and server components |website|link | demo|
|
spsComps | SPS UI and server components |website|link | demo|
|![]() drawer | SPS interactive image editing tool |website|link | demo|
|
drawer | SPS interactive image editing tool |website|link | demo|
|![]() spsUtil | SPS utility functions |website|link | NA|
spsUtil | SPS utility functions |website|link | NA|
More Tutorials
ExpandYou need to unmuted it to hear the sound.
Workflow Module
RNAseq Module
Quick ggplot Module
Canvas Tool
Screenshots of SPS
ExpandFull app

Loading screens

Workflow module

Workflow Execution

RNASeq module


Canvas

Admin



Debugging

Contact & contributions
Please use https://github.com/systemPipeR/systemPipeShiny/issues for reporting bugs, issues or for suggesting new features to be implemented.
We'd love to hear from all users and developers. Submit your pull request if you have new thoughts or improvements.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
