script/mutation_detection.md
In zhouzilu/DENDRO: Dna based EvolutioNary tree preDiction by scRna-seq technOlogy
DENDRO mutation detection with GATK
DENDRO uses the following approach in the original paper to generate the initial mutation profiles across genes and cells. We write an example script modified from one real data analysis. Just want to clarify that I am not an expert with GATK tool. However, given significant amount of tests and failures, the following pipeline works for DENDRO. See HaplotypeCaller document for more information.
Questions, Suggestions & Problems
If you have any questions or problems when using DENDRO or DENDROplan, please feel free to open a new issue here. You can also email the maintainers of the corresponding packages -- the contact information is shown under Developers & Maintainers.
Pipeline overview
Initial SNA detection is indeed one of the most important steps in DENDRO pipeline. As stated in the paper, we want to maximize the sensitivity of our detection at this moment. There will be a lot of false positives calls, but they will be cleaned up in the clustering and pooling steps. Figure 4 shows the detection pipeline. User should check the example script together with this figure side-by-side.
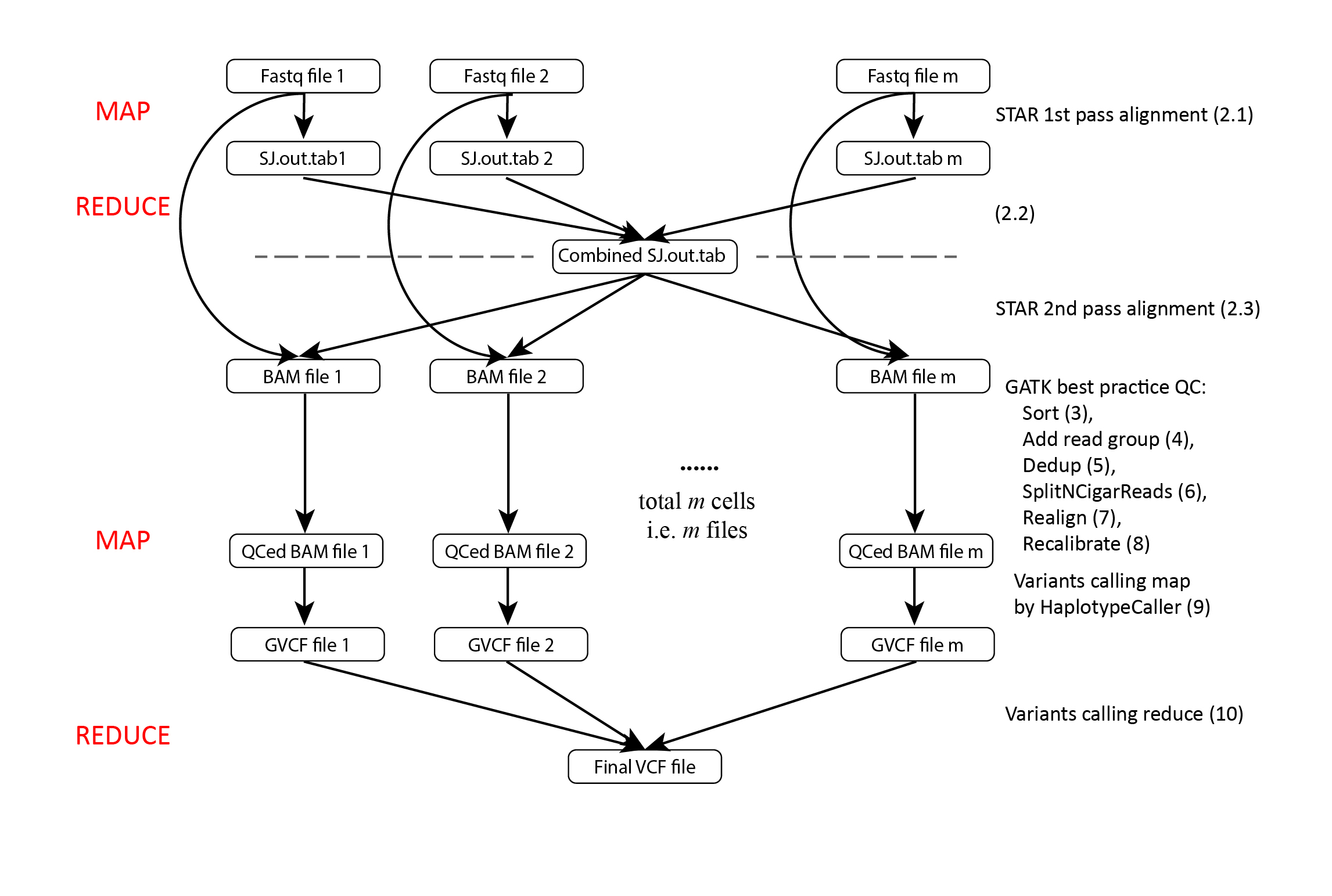
Figure 5. A flowchart outlining the procedures of mutation detection from fastq to final GVCF using GATK ERC GVCF approach. The number on the most right corresponding to the steps in the shell script.
Due to great number of cells, traditional way of calling variants sample by sample is extremely slow. Luckily, GATK has ERC GVCF mode, which utilize a map-reduce like approach. Please check the details here. Our script is built upon ERC GVCF mode togehter with the variant detection best approach on RNAseq data.
Example script is attached here. We also provided an R script to convert vcf files to DENDRO input.
Citation
Zhou, Z., Xu, B., Minn, A. et al. DENDRO: genetic heterogeneity profiling and subclone detection by single-cell RNA sequencing. Genome Biol 21, 10 (2020). https://doi.org/10.1186/s13059-019-1922-x
Developers & Maintainers
-
Zilu Zhou (zhouzilu at pennmedicine dot upenn dot edu)
Genomics and Computational Biology Graduate Group, University of Pennsylvania
-
Nancy R. Zhang
Department of Statistics, University of Pennsylvania
zhouzilu/DENDRO documentation built on May 21, 2020, 8 p.m.
DENDRO mutation detection with GATK
DENDRO uses the following approach in the original paper to generate the initial mutation profiles across genes and cells. We write an example script modified from one real data analysis. Just want to clarify that I am not an expert with GATK tool. However, given significant amount of tests and failures, the following pipeline works for DENDRO. See HaplotypeCaller document for more information.
Questions, Suggestions & Problems
If you have any questions or problems when using DENDRO or DENDROplan, please feel free to open a new issue here. You can also email the maintainers of the corresponding packages -- the contact information is shown under Developers & Maintainers.
Pipeline overview
Initial SNA detection is indeed one of the most important steps in DENDRO pipeline. As stated in the paper, we want to maximize the sensitivity of our detection at this moment. There will be a lot of false positives calls, but they will be cleaned up in the clustering and pooling steps. Figure 4 shows the detection pipeline. User should check the example script together with this figure side-by-side.

Figure 5. A flowchart outlining the procedures of mutation detection from fastq to final GVCF using GATK ERC GVCF approach. The number on the most right corresponding to the steps in the shell script.
Due to great number of cells, traditional way of calling variants sample by sample is extremely slow. Luckily, GATK has ERC GVCF mode, which utilize a map-reduce like approach. Please check the details here. Our script is built upon ERC GVCF mode togehter with the variant detection best approach on RNAseq data.
Example script is attached here. We also provided an R script to convert vcf files to DENDRO input.
Citation
Zhou, Z., Xu, B., Minn, A. et al. DENDRO: genetic heterogeneity profiling and subclone detection by single-cell RNA sequencing. Genome Biol 21, 10 (2020). https://doi.org/10.1186/s13059-019-1922-x
Developers & Maintainers
-
Zilu Zhou (zhouzilu at pennmedicine dot upenn dot edu) Genomics and Computational Biology Graduate Group, University of Pennsylvania
-
Nancy R. Zhang Department of Statistics, University of Pennsylvania
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
