README.md
In TreeLS: Terrestrial Point Cloud Processing of Forest Data
TreeLS
High performance R functions for forest data processing based on Terrestrial Laser Scanning (but not only) point clouds.
Description
This package is a refactor of the methods described in this paper, among many other features for 3D point cloud processing of forest environments.
Most algorithms are written in C++ and wrapped in R functions through Rcpp. TreeLS is built on top of lidR, using its LAS infrastructure internally for most methods.
For any questions, comments or bug reports please submit an issue here on GitHub. Suggestions, ideas and references of new algorithms are always welcome - as long as they fit into TreeLS' scope.
TreeLS is currently on v2.0. To install it from an official mirror, use: install.packages("TreeLS"). To install the most recent version, check out the Installation from source section below.
*TreeLS is not on CRAN at the moment (August/2020), the up-to-date version is submitted and should be available shortly. Meanwhile you can install it from source using devtools.
News
-
August/2020: Version 2.0 is finally available! It's a major release, introducing several new functionalities, bug fixes, more robust estimators for noisy clouds and more flexible plotting. All functionalities from older versions are now available and optimized, so there should be no need to use legacy code anymore. The scope of application of TreeLS has become much wider in this version, specially due to the introduction of functions like fastPointMetrics and shapeFit, making it much easier for researchers to assess point cloud data in many contexts and develop their own methods on top of those functions. For a comprehensive list of the updates check out the CHANGELOG.
-
March/2019: TreeLS is finally available on CRAN and is now an official R package.
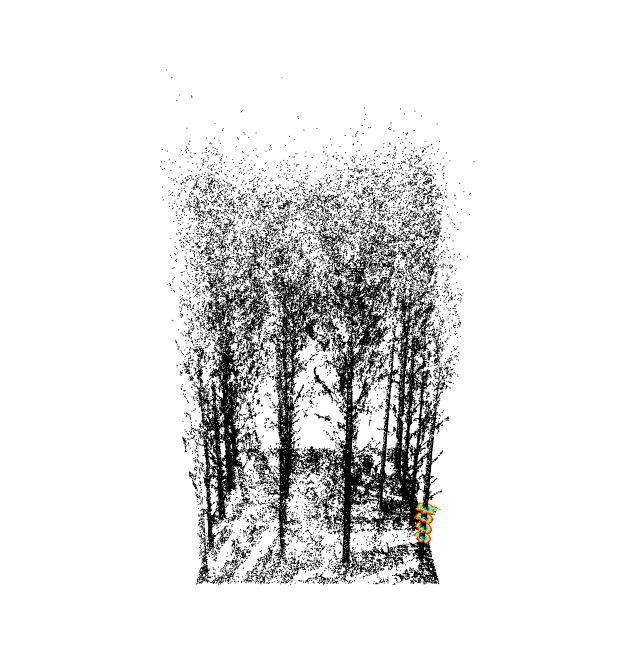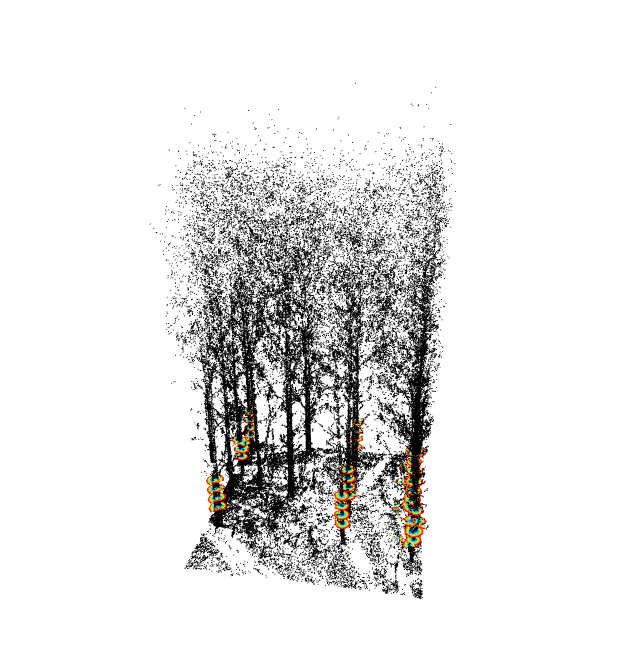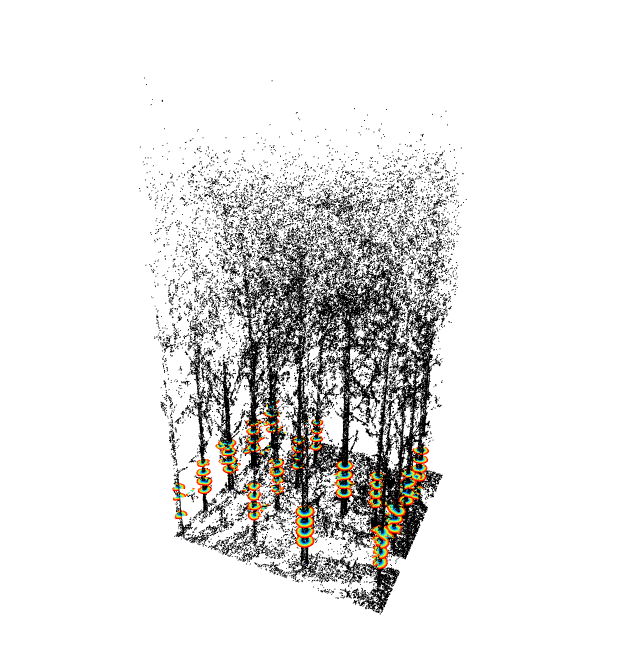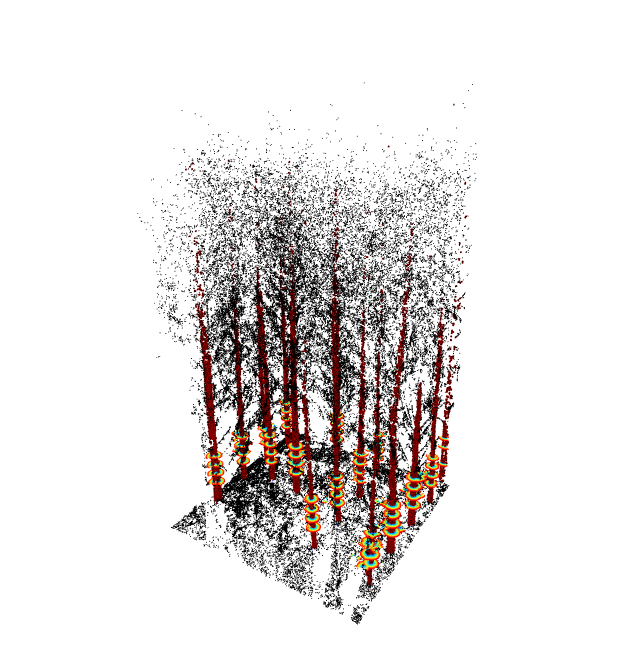
Main functionalities
- Tree detection at plot level
- Tree region assignment
- Stem detection and denoising
- Stem segmentation
- Forest inventory
- Fast calculation of point features
- Research basis and other applications
- 3D plotting and manipulation
Installation from source
Requirements
devtools: run install.packages('devtools', dependencies = TRUE) from the R console- Rcpp compiler:
- on Windows: install Rtools for your R version - make sure to add it to your system's path
- on Mac: install Xcode
- on Linux: be sure to have
r-base-dev installed
Install TreeLS latest version
On the R console, run:
devtools::install_github('tiagodc/TreeLS')
Usage
Example of full processing workflow from reading a point cloud file until stem segmentation of a forest plot:
library(TreeLS)
# open sample plot file
file = system.file("extdata", "pine_plot.laz", package="TreeLS")
tls = readTLS(file)
# normalize the point cloud
tls = tlsNormalize(tls, keep_ground = F)
x = plot(tls)
# extract the tree map from a thinned point cloud
thin = tlsSample(tls, smp.voxelize(0.02))
map = treeMap(thin, map.hough(min_density = 0.1), 0)
add_treeMap(x, map, color='yellow', size=2)
# classify tree regions
tls = treePoints(tls, map, trp.crop())
add_treePoints(x, tls, size=4)
add_treeIDs(x, tls, cex = 2, col='yellow')
# classify stem points
tls = stemPoints(tls, stm.hough())
add_stemPoints(x, tls, color='red', size=8)
# make the plot's inventory
inv = tlsInventory(tls, d_method=shapeFit(shape='circle', algorithm = 'irls'))
add_tlsInventory(x, inv)
# extract stem measures
seg = stemSegmentation(tls, sgt.ransac.circle(n = 20))
add_stemSegments(x, seg, color='white', fast=T)
# plot everything once
tlsPlot(tls, map, inv, seg, fast=T)
# check out only one tree
tlsPlot(tls, inv, seg, tree_id = 11)
#------------------------------------------#
### overview of some new methods on v2.0 ###
#------------------------------------------#
file = system.file("extdata", "pine.laz", package="TreeLS")
tls = readTLS(file) %>% tlsNormalize()
# calculate some point metrics
tls = fastPointMetrics(tls, ptm.knn())
x = plot(tls, color='Verticality')
# get its stem points
tls = stemPoints(tls, stm.eigen.knn(voxel_spacing = .02))
add_stemPoints(x, tls, size=3, color='red')
# get dbh and height
dbh_algo = shapeFit(shape='cylinder', algorithm = 'bf', n=15, inliers=.95, z_dev=10)
inv = tlsInventory(tls, hp = .95, d_method = dbh_algo)
add_tlsInventory(x, inv)
# segment the stem usind 3D cylinders and getting their directions
seg = stemSegmentation(tls, sgt.irls.cylinder(n=300))
add_stemSegments(x, seg, color='blue')
# check out a specific tree segment
tlsPlot(seg, tls, segment = 3)
Try the TreeLS package in your browser
Any scripts or data that you put into this service are public.
TreeLS documentation built on Aug. 26, 2020, 5:14 p.m.
TreeLS
High performance R functions for forest data processing based on Terrestrial Laser Scanning (but not only) point clouds.
Description
This package is a refactor of the methods described in this paper, among many other features for 3D point cloud processing of forest environments.
Most algorithms are written in C++ and wrapped in R functions through Rcpp. TreeLS is built on top of lidR, using its LAS infrastructure internally for most methods.
For any questions, comments or bug reports please submit an issue here on GitHub. Suggestions, ideas and references of new algorithms are always welcome - as long as they fit into TreeLS' scope.
TreeLS is currently on v2.0. To install it from an official mirror, use: install.packages("TreeLS"). To install the most recent version, check out the Installation from source section below.
*TreeLS is not on CRAN at the moment (August/2020), the up-to-date version is submitted and should be available shortly. Meanwhile you can install it from source using devtools.
News
-
August/2020: Version 2.0 is finally available! It's a major release, introducing several new functionalities, bug fixes, more robust estimators for noisy clouds and more flexible plotting. All functionalities from older versions are now available and optimized, so there should be no need to use legacy code anymore. The scope of application of TreeLS has become much wider in this version, specially due to the introduction of functions like
fastPointMetricsandshapeFit, making it much easier for researchers to assess point cloud data in many contexts and develop their own methods on top of those functions. For a comprehensive list of the updates check out the CHANGELOG. -
March/2019:
TreeLSis finally available on CRAN and is now an official R package.
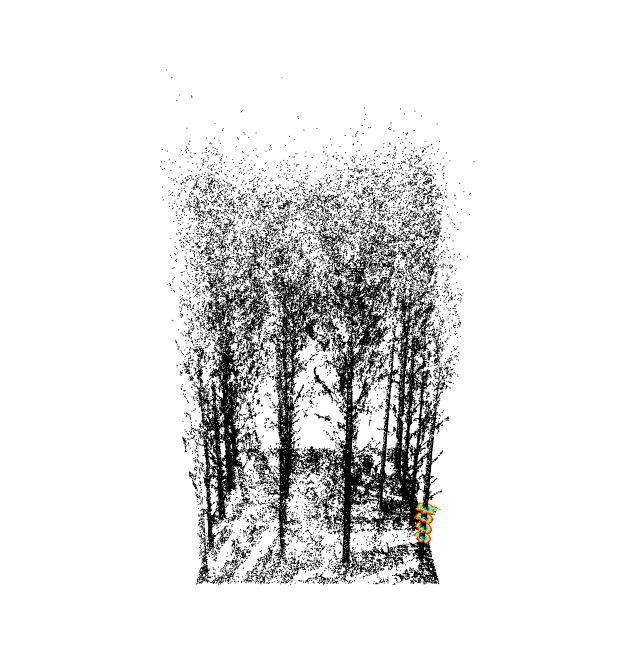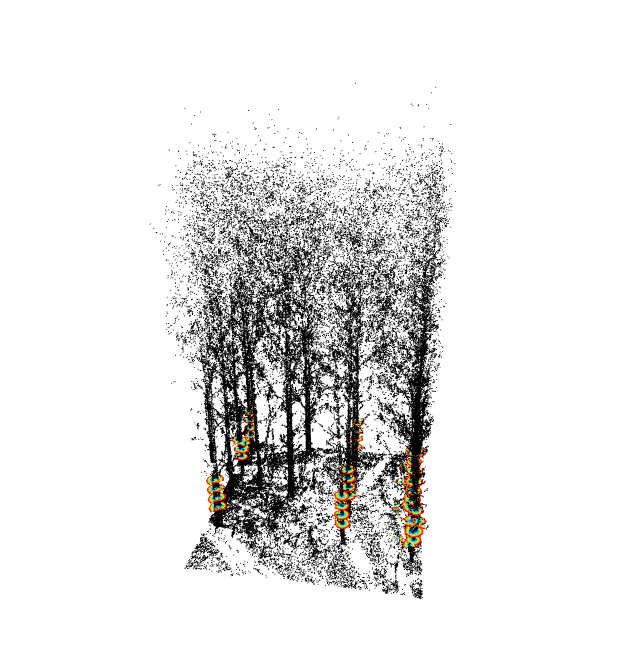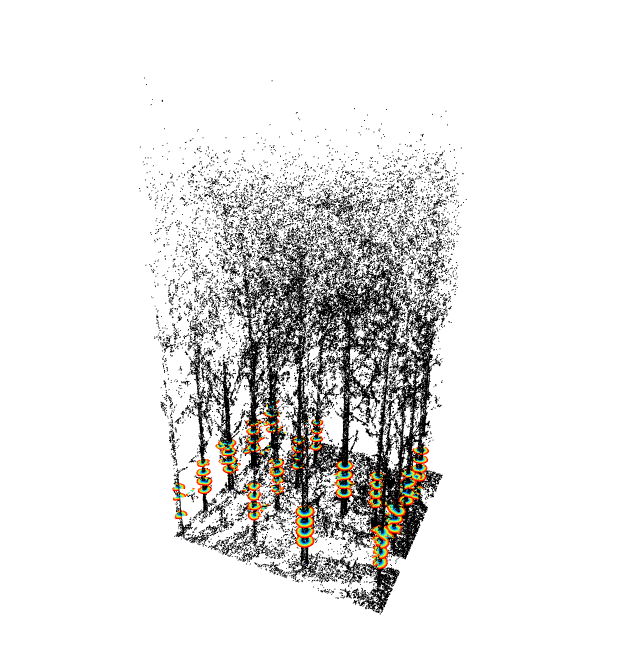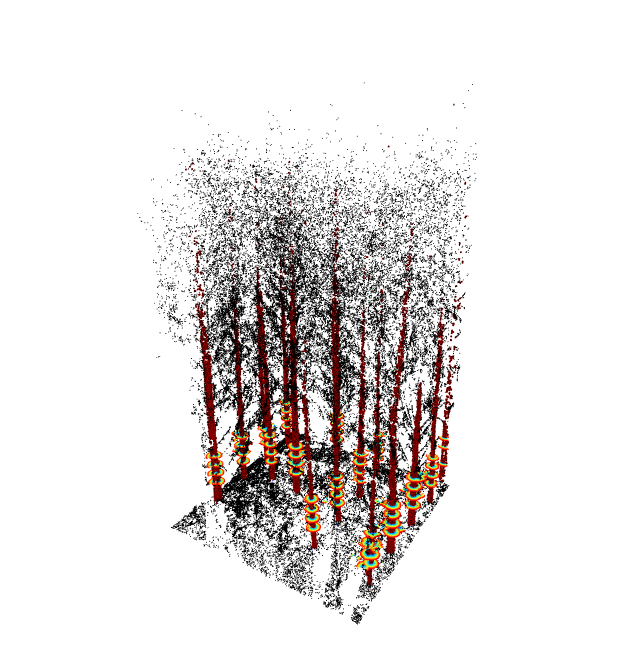
Main functionalities
- Tree detection at plot level
- Tree region assignment
- Stem detection and denoising
- Stem segmentation
- Forest inventory
- Fast calculation of point features
- Research basis and other applications
- 3D plotting and manipulation
Installation from source
Requirements
devtools: runinstall.packages('devtools', dependencies = TRUE)from the R console- Rcpp compiler:
- on Windows: install Rtools for your R version - make sure to add it to your system's path
- on Mac: install Xcode
- on Linux: be sure to have
r-base-devinstalled
Install TreeLS latest version
On the R console, run:
devtools::install_github('tiagodc/TreeLS')
Usage
Example of full processing workflow from reading a point cloud file until stem segmentation of a forest plot:
library(TreeLS)
# open sample plot file
file = system.file("extdata", "pine_plot.laz", package="TreeLS")
tls = readTLS(file)
# normalize the point cloud
tls = tlsNormalize(tls, keep_ground = F)
x = plot(tls)
# extract the tree map from a thinned point cloud
thin = tlsSample(tls, smp.voxelize(0.02))
map = treeMap(thin, map.hough(min_density = 0.1), 0)
add_treeMap(x, map, color='yellow', size=2)
# classify tree regions
tls = treePoints(tls, map, trp.crop())
add_treePoints(x, tls, size=4)
add_treeIDs(x, tls, cex = 2, col='yellow')
# classify stem points
tls = stemPoints(tls, stm.hough())
add_stemPoints(x, tls, color='red', size=8)
# make the plot's inventory
inv = tlsInventory(tls, d_method=shapeFit(shape='circle', algorithm = 'irls'))
add_tlsInventory(x, inv)
# extract stem measures
seg = stemSegmentation(tls, sgt.ransac.circle(n = 20))
add_stemSegments(x, seg, color='white', fast=T)
# plot everything once
tlsPlot(tls, map, inv, seg, fast=T)
# check out only one tree
tlsPlot(tls, inv, seg, tree_id = 11)
#------------------------------------------#
### overview of some new methods on v2.0 ###
#------------------------------------------#
file = system.file("extdata", "pine.laz", package="TreeLS")
tls = readTLS(file) %>% tlsNormalize()
# calculate some point metrics
tls = fastPointMetrics(tls, ptm.knn())
x = plot(tls, color='Verticality')
# get its stem points
tls = stemPoints(tls, stm.eigen.knn(voxel_spacing = .02))
add_stemPoints(x, tls, size=3, color='red')
# get dbh and height
dbh_algo = shapeFit(shape='cylinder', algorithm = 'bf', n=15, inliers=.95, z_dev=10)
inv = tlsInventory(tls, hp = .95, d_method = dbh_algo)
add_tlsInventory(x, inv)
# segment the stem usind 3D cylinders and getting their directions
seg = stemSegmentation(tls, sgt.irls.cylinder(n=300))
add_stemSegments(x, seg, color='blue')
# check out a specific tree segment
tlsPlot(seg, tls, segment = 3)
Try the TreeLS package in your browser
Any scripts or data that you put into this service are public.
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.


