README.md
In brentp/celltypes450: Deal with cell types in 450K data.
Adjust for cell composition in 450K data.
Much of the adjust.beta code was originally written by Andres Houseman.
I have only put it into a simple to use function and packaged it.
Installation
> library(devtools)
> install_github("brentp/celltypes450")
About
See this thread for details.
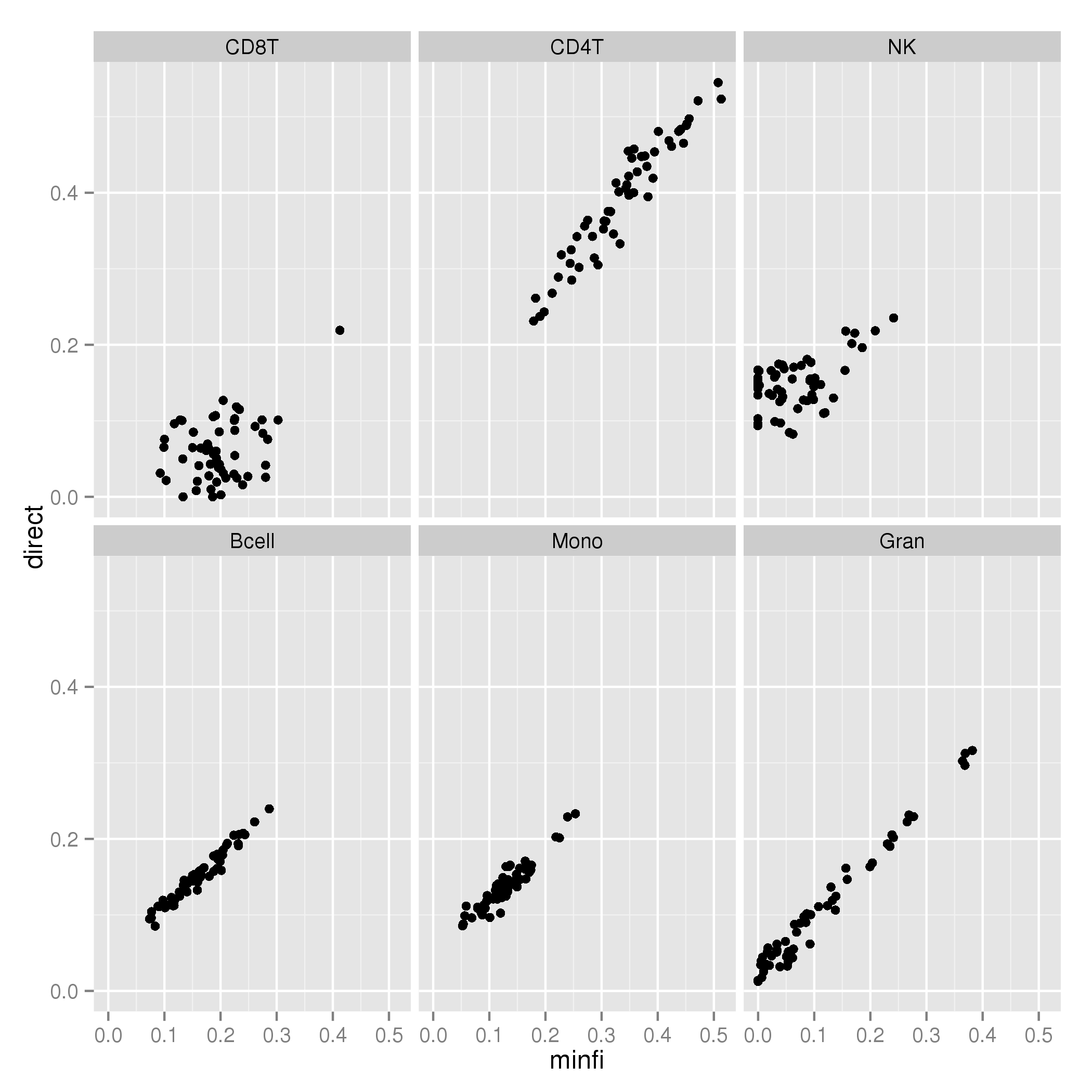
Compared to minfi, which normalizes the sorted data with the new data:
To get your beta (0, 1) values adjusted for cell-type composition, use:
library(devtools)
install_github("brentp/celltypes450")
library(celltypes450)
adjusted = adjust.beta(beta)
To get the cell-composition estimates such as those from the image above:
cell.types = adjust.beta(beta, est.only=TRUE)
brentp/celltypes450 documentation built on May 13, 2019, 5:11 a.m.
Adjust for cell composition in 450K data.
Much of the adjust.beta code was originally written by Andres Houseman. I have only put it into a simple to use function and packaged it.
Installation
> library(devtools)
> install_github("brentp/celltypes450")
About
See this thread for details.
Compared to minfi, which normalizes the sorted data with the new data:

To get your beta (0, 1) values adjusted for cell-type composition, use:
library(devtools)
install_github("brentp/celltypes450")
library(celltypes450)
adjusted = adjust.beta(beta)
To get the cell-composition estimates such as those from the image above:
cell.types = adjust.beta(beta, est.only=TRUE)
Embedding an R snippet on your website
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
