inst/examples/labrid_data.md
In cboettig/wrightscape: wrightscape
labrid data processing
require(phytools)
require(geiger)
require(wrightscape)
(This data has not been released)
labrid_tree <- read.nexus("../../data/labrid_tree.nex")
#fin_data <-read.csv(paste(path,"labrid.csv", sep="")) # not actually being used
diet_data <- read.csv("../../data/labriddata_parrotfish.csv")
Simple size corrections length and weight as fraction of body mass could use
for(i in c(3,4,6,7,8)){
diet_data[i] <- diet_data[i]/diet_data[5]
}
diet_data[5] <- log(diet_data[5])
We will take a different approach, using the phylogenetic correction, so the above code is not to be run.
corrected_data <- diet_data
Use the simple trait names from @Price2010
traitnames <- c("Species", "group", "gape", "prot", "bodymass", "AM", "SH", "LP", "close", "open", "kt")
names(corrected_data) <- traitnames
Lengths are log transformed
corrected_data[["gape"]] <- log(corrected_data[["gape"]])
corrected_data[["prot"]] <- log(corrected_data[["prot"]])
masses are log(cube-root) transformed
corrected_data[["bodymass"]] <- log(corrected_data[["bodymass"]])/3
corrected_data[["AM"]] <- log(corrected_data[["AM"]])/3
corrected_data[["SH"]] <- log(corrected_data[["SH"]])/3
corrected_data[["LP"]] <- log(corrected_data[["LP"]])/3
Traits which are ratios are fine as they are
Drop any unmatched tip-traits
ape <- treedata(labrid_tree, corrected_data[,3:11], corrected_data[,1])
Dropped rows from the data because there were no matching tips in the tree:
[1] Calotomus_carolinus Coris_aurilineata
[3] Halichoeres_melasmapomus Leptoscarus_vaigiensis
[5] Scarus_forsteni Scarus_quoyi
[7] Scarus_rubroviolaceus Xiphocheilus_typus
122 Levels: Anampses_caeruleopunctatus ... Xyrichtys_novacula
Run Revell's phylogenetic size corrections
ape$data["bodysize"]
[1] NA
out <- phyl.resid(ape$phy, ape$data[,"bodymass"], ape$data[,c("gape", "prot","AM", "SH", "LP")] )
The phyl.resid function changes order of species listing. Merge for a set of uncorrected and corrected traits.
traits <- merge(ape$data, out$resid, by="row.names")
columns that are transformed now have gape.x for untransformed, gape.y for transformed.
format_data() gets regimes from column specified in
"regimes" (e.g. this is a column id, not a # of regimes)
This also converts the tree and data into ouch format
(We could just hand it all traits, but these are just the tranformed and size-corrected ones)
labrid <- format_data(labrid_tree, traits[,2:length(traits)], species_names=traits[,1])
Painting Regimes
Select common ancestor of a Chlorurus and a Hipposcarus as the changepoint
intra_ancestor <- mrcaOUCH(c("Chlorurus_sordidus", "Hipposcarus_longiceps"), labrid$tree)
intramandibular <- paintBranches(intra_ancestor, labrid$tree, c("other","intramandibular"))
Select common ancestor for all parrot fish:
pharyngeal_ancestor <- mrcaOUCH(c("Bolbometopon_muricatum", "Sparisoma_radians"), labrid$tree)
pharyngeal <- paintBranches(pharyngeal_ancestor, labrid$tree, c("other","pharyngeal"))
two_shifts <- paintBranches(c(pharyngeal_ancestor, intra_ancestor), labrid$tree, c("wrasses", "pharyngeal", "intramandibular") )
This leaves the branch on which the second transition occurs unspecified (fourth regime).
We have to fix this manually
two_shifts[as.numeric(intra_ancestor)] <- "intramandibular"
two_shifts <- as.factor(as.character(two_shifts))
names(two_shifts) <- names(intramandibular)
rename the ouch-formated data
dat <- labrid$data
tree <- labrid$tree
rename the ape-formatted data
ape.phy <- ape$phy
ape.dat <- traits
Save the final output. (This data is provided with the package).
save(list=c("intramandibular", "pharyngeal", "two_shifts", "tree", "dat", "ape.phy", "ape.dat"), file="labrids.rda")
We now have access to the following configurations:
intramandibular, pharyngeal and two_shifts paintings, (and labrid$noregimes),
and tree in labrid$tree
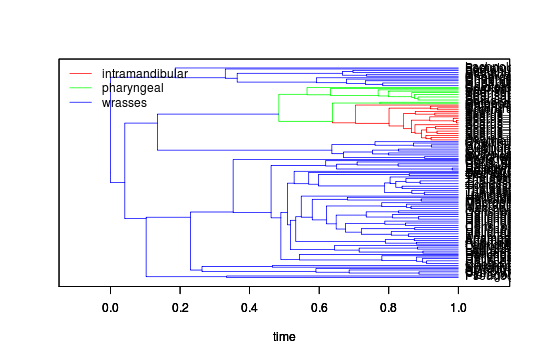
plot(labrid$tree, regimes=two_shifts)
cboettig/wrightscape documentation built on May 13, 2019, 2:12 p.m.
labrid data processing
require(phytools)
require(geiger)
require(wrightscape)
(This data has not been released)
labrid_tree <- read.nexus("../../data/labrid_tree.nex")
#fin_data <-read.csv(paste(path,"labrid.csv", sep="")) # not actually being used
diet_data <- read.csv("../../data/labriddata_parrotfish.csv")
Simple size corrections length and weight as fraction of body mass could use
for(i in c(3,4,6,7,8)){
diet_data[i] <- diet_data[i]/diet_data[5]
}
diet_data[5] <- log(diet_data[5])
We will take a different approach, using the phylogenetic correction, so the above code is not to be run.
corrected_data <- diet_data
Use the simple trait names from @Price2010
traitnames <- c("Species", "group", "gape", "prot", "bodymass", "AM", "SH", "LP", "close", "open", "kt")
names(corrected_data) <- traitnames
Lengths are log transformed
corrected_data[["gape"]] <- log(corrected_data[["gape"]])
corrected_data[["prot"]] <- log(corrected_data[["prot"]])
masses are log(cube-root) transformed
corrected_data[["bodymass"]] <- log(corrected_data[["bodymass"]])/3
corrected_data[["AM"]] <- log(corrected_data[["AM"]])/3
corrected_data[["SH"]] <- log(corrected_data[["SH"]])/3
corrected_data[["LP"]] <- log(corrected_data[["LP"]])/3
Traits which are ratios are fine as they are
Drop any unmatched tip-traits
ape <- treedata(labrid_tree, corrected_data[,3:11], corrected_data[,1])
Dropped rows from the data because there were no matching tips in the tree:
[1] Calotomus_carolinus Coris_aurilineata
[3] Halichoeres_melasmapomus Leptoscarus_vaigiensis
[5] Scarus_forsteni Scarus_quoyi
[7] Scarus_rubroviolaceus Xiphocheilus_typus
122 Levels: Anampses_caeruleopunctatus ... Xyrichtys_novacula
Run Revell's phylogenetic size corrections
ape$data["bodysize"]
[1] NA
out <- phyl.resid(ape$phy, ape$data[,"bodymass"], ape$data[,c("gape", "prot","AM", "SH", "LP")] )
The phyl.resid function changes order of species listing. Merge for a set of uncorrected and corrected traits.
traits <- merge(ape$data, out$resid, by="row.names")
columns that are transformed now have gape.x for untransformed, gape.y for transformed.
format_data() gets regimes from column specified in
"regimes" (e.g. this is a column id, not a # of regimes)
This also converts the tree and data into ouch format
(We could just hand it all traits, but these are just the tranformed and size-corrected ones)
labrid <- format_data(labrid_tree, traits[,2:length(traits)], species_names=traits[,1])
Painting Regimes
Select common ancestor of a Chlorurus and a Hipposcarus as the changepoint
intra_ancestor <- mrcaOUCH(c("Chlorurus_sordidus", "Hipposcarus_longiceps"), labrid$tree)
intramandibular <- paintBranches(intra_ancestor, labrid$tree, c("other","intramandibular"))
Select common ancestor for all parrot fish:
pharyngeal_ancestor <- mrcaOUCH(c("Bolbometopon_muricatum", "Sparisoma_radians"), labrid$tree)
pharyngeal <- paintBranches(pharyngeal_ancestor, labrid$tree, c("other","pharyngeal"))
two_shifts <- paintBranches(c(pharyngeal_ancestor, intra_ancestor), labrid$tree, c("wrasses", "pharyngeal", "intramandibular") )
This leaves the branch on which the second transition occurs unspecified (fourth regime). We have to fix this manually
two_shifts[as.numeric(intra_ancestor)] <- "intramandibular"
two_shifts <- as.factor(as.character(two_shifts))
names(two_shifts) <- names(intramandibular)
rename the ouch-formated data
dat <- labrid$data
tree <- labrid$tree
rename the ape-formatted data
ape.phy <- ape$phy
ape.dat <- traits
Save the final output. (This data is provided with the package).
save(list=c("intramandibular", "pharyngeal", "two_shifts", "tree", "dat", "ape.phy", "ape.dat"), file="labrids.rda")
We now have access to the following configurations:
intramandibular, pharyngeal and two_shifts paintings, (and labrid$noregimes),
and tree in labrid$tree
plot(labrid$tree, regimes=two_shifts)

Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
