inst/examples/labrid_mc.md
In cboettig/wrightscape: wrightscape
- Author: Carl Boettiger cboettig@gmail.com
- License: BSD
require(wrightscape)
require(snowfall)
require(ggplot2)
require(reshape)
data(labrids)
traits <- c("bodymass", "close", "open", "kt", "gape.y", "prot.y", "AM.y", "SH.y", "LP.y")
regimes <- two_shifts
Just a few processors, for debugging locally.
sfInit(par=T, 4)
sfLibrary(wrightscape)
Library wrightscape loaded.
sfExportAll()
The main parallel loop fitting each model
fits <- sfLapply(traits, function(trait){
multi <- function(modelspec){
out <- multiTypeOU(data = dat[[trait]], tree = tree, regimes = regimes,
model_spec = modelspec, control = list(maxit=8000))
n <- length(levels(out$regimes))
Xo <- rep(out$Xo,n)
loglik <- rep(out$loglik, n)
pars <- cbind(out$alpha, out$sigma, out$theta, Xo, loglik)
rownames(pars) <- levels(out$regimes)
colnames(pars) <- c("alpha", "sigma", "theta", "Xo", "loglik")
if(out$convergence != 0) # only return values if successful
pars[,] <- NA
pars
}
bm <- multi(list(alpha = "fixed", sigma = "global", theta = "global"))
ou <- multi(list(alpha = "global", sigma = "global", theta = "global"))
bm2 <- multi(list(alpha = "fixed", sigma = "indep", theta = "global"))
a2 <- multi(list(alpha = "indep", sigma = "global", theta = "global"))
t2 <- multi(list(alpha = "global", sigma = "global", theta = "indep"))
list(bm=bm,brownie=bm2, ou=ou, ouch=t2, alphas=a2)
})
Reformat and label data for plotting
names(fits) <- traits # each fit is a different trait (so use it for a label)
data <- melt(fits)
names(data) <- c("regimes", "param", "value", "model", "trait")
model likelihood
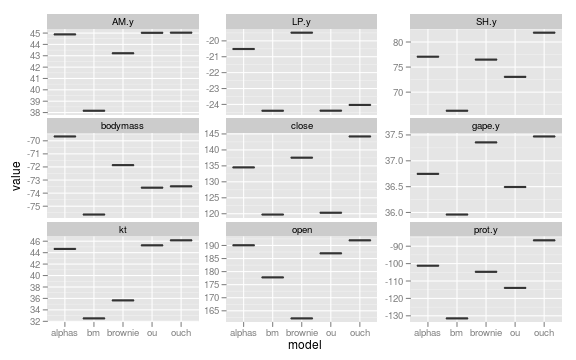
ggplot(subset(data, param=="loglik")) +
geom_boxplot(aes(model, value)) +
facet_wrap(~ trait, scales="free_y")
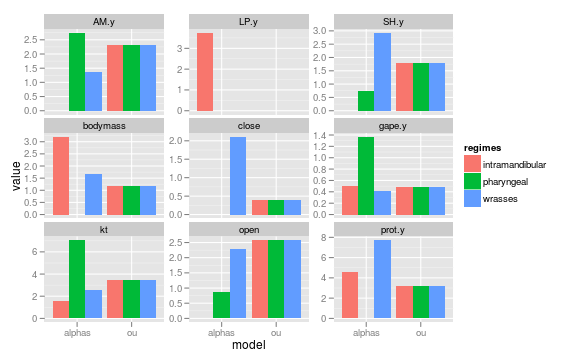
Parameter distributions of alpha parameter in model alpha (alphas vary) and ou (global).
ggplot(subset(data, param %in% c("alpha")
& model %in% c("alphas", "ou")),
aes(model, value, fill=regimes)) +
geom_bar(position="dodge") +
facet_wrap(~trait, scales="free_y")
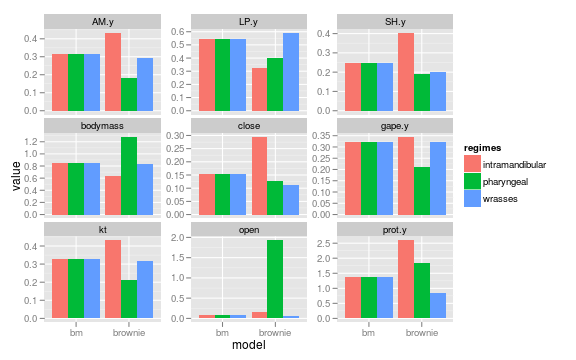
Parameter distribution of the sigma parameter in the brownie and bm models
ggplot(subset(data, param %in% c("sigma")
& model %in% c("bm", "brownie")),
aes(model, value, fill=regimes)) +
geom_bar(position="dodge") +
facet_wrap(~trait, scales="free_y")
save(list=ls(), file="~/public_html/data/labrid_mc.rda")
cboettig/wrightscape documentation built on May 13, 2019, 2:12 p.m.
- Author: Carl Boettiger cboettig@gmail.com
- License: BSD
require(wrightscape)
require(snowfall)
require(ggplot2)
require(reshape)
data(labrids)
traits <- c("bodymass", "close", "open", "kt", "gape.y", "prot.y", "AM.y", "SH.y", "LP.y")
regimes <- two_shifts
Just a few processors, for debugging locally.
sfInit(par=T, 4)
sfLibrary(wrightscape)
Library wrightscape loaded.
sfExportAll()
The main parallel loop fitting each model
fits <- sfLapply(traits, function(trait){
multi <- function(modelspec){
out <- multiTypeOU(data = dat[[trait]], tree = tree, regimes = regimes,
model_spec = modelspec, control = list(maxit=8000))
n <- length(levels(out$regimes))
Xo <- rep(out$Xo,n)
loglik <- rep(out$loglik, n)
pars <- cbind(out$alpha, out$sigma, out$theta, Xo, loglik)
rownames(pars) <- levels(out$regimes)
colnames(pars) <- c("alpha", "sigma", "theta", "Xo", "loglik")
if(out$convergence != 0) # only return values if successful
pars[,] <- NA
pars
}
bm <- multi(list(alpha = "fixed", sigma = "global", theta = "global"))
ou <- multi(list(alpha = "global", sigma = "global", theta = "global"))
bm2 <- multi(list(alpha = "fixed", sigma = "indep", theta = "global"))
a2 <- multi(list(alpha = "indep", sigma = "global", theta = "global"))
t2 <- multi(list(alpha = "global", sigma = "global", theta = "indep"))
list(bm=bm,brownie=bm2, ou=ou, ouch=t2, alphas=a2)
})
Reformat and label data for plotting
names(fits) <- traits # each fit is a different trait (so use it for a label)
data <- melt(fits)
names(data) <- c("regimes", "param", "value", "model", "trait")
model likelihood
ggplot(subset(data, param=="loglik")) +
geom_boxplot(aes(model, value)) +
facet_wrap(~ trait, scales="free_y")

Parameter distributions of alpha parameter in model alpha (alphas vary) and ou (global).
ggplot(subset(data, param %in% c("alpha")
& model %in% c("alphas", "ou")),
aes(model, value, fill=regimes)) +
geom_bar(position="dodge") +
facet_wrap(~trait, scales="free_y")

Parameter distribution of the sigma parameter in the brownie and bm models
ggplot(subset(data, param %in% c("sigma")
& model %in% c("bm", "brownie")),
aes(model, value, fill=regimes)) +
geom_bar(position="dodge") +
facet_wrap(~trait, scales="free_y")

save(list=ls(), file="~/public_html/data/labrid_mc.rda")
Embedding an R snippet on your website
Add the following code to your website.
For more information on customizing the embed code, read Embedding Snippets.
